Research Article - Biomedical Research (2017) Volume 28, Issue 18
XRCC1 gene rs25487, rs1799782 polymorphisms do not influence the susceptibility of CAD
Ying-Nan Wang, De-Wei Wang, Lei Gao, Shi-Yong Wu, Dong-Yin Zhang and Shu Qin*
Department of Cardiology, the First Affiliated Hospital of Chongqing Medical University, Chongqing, PR China
- *Corresponding Author:
- Shu Qin
Department of Cardiology
The First Affiliated Hospital of Chongqing Medical University, PR China
Accepted on August 21, 2017
Abstract
Background: Two meta-analyses assessed the association between X-Ray Repair Cross Complementing 1 (XRCC1) gene polymorphisms and Coronary Artery Disease (CAD) risk. However, these two metaanalyses all included the studies without Hardy-Weinberg equilibrium (HWE). Therefore, their results were not reliable.
Methods: PubMed, EMBASE, MEDLINE, and the Cochrane Library were searched up to May 2017. Odds Ratios (OR) and 95% Confidence Interval (CI) were evaluated to determine the strength.
Results: Five case-control studies with 1715 cases and 1520 controls were included in this meta-analysis. A significant association was not found between XRCC1 rs25487 polymorphism and CAD risk (OR=0.93; 95% CI, 0.72-1.21; P=0.61). No significant association was found between XRCC1 rs25487 polymorphism and CAD risk in Caucasian (OR=0.93; 95% CI, 0.63-1.39; P=0.74) and Asian (OR=0.93; 95% CI, 0.50-1.75; P=0.83). A significant association was also not found between XRCC1 rs1799782 polymorphism and CAD risk (OR=1.07; 95% CI, 0.92-1.25; P=0.39). In the Caucasian subgroup, no significant association was observed (OR=1.07; 95% CI, 0.92-1.25; P=0.39).
Conclusions: This meta-analysis suggested that XRCC1 gene rs25487, rs1799782 polymorphisms were not associated with CAD risk.
Keywords
XRCC1, CAD, Polymorphism.
Introduction
Coronary Artery Disease (CAD) is very common with a prevalence of 6.9% in males and 6% in females [1]. It is a multifactorial disease. Many reports suggested that this disease is resulting from interactions between environmental and genetic factors [2]. More recently, some studies indicated the role of genetic factor in CAD development [3].
The protein of X-ray Repair Cross Complementing 1 (XRCC1) play an important role in diseases development [4]. XRCC1 gene polymorphisms were associated with disease susceptibility. Forat-Yazdi et al. found that Arg399Gln in XRCC1 was associated with colorectal cancer risk [5]. Yan et al. suggested that this polymorphism was associated with the risk of prostate cancer [6]. Fan et al. suggested that the XRCC1 Arg194Trp polymorphism was associated with the efficacy of platinum-based chemotherapy [7]. Sanjari et al. suggested that XRCC1 gene polymorphisms were associated with breast cancer risk [8]. Previous studies have investigated the association between XRCC1 gene polymorphisms and CAD risk [9-13]. Two meta-analyses also tried to determine the association between XRCC1 gene polymorphisms and CAD risk [14,15]. Guo et al. suggested that the Arg194Trp polymorphism contributed to the CAD risk [14]. Ma et al. suggested that the XRCC1 gene Arg194Trp and Arg399Gln polymorphisms were associated with CAD susceptibility [15]. However, these two meta-analysis all included the studies without Hardy-Weinberg Equilibrium (HWE). Therefore, the results were not reliable.
Materials and Methods
Publications search
PubMed, EMBASE, MEDLINE, and the Cochrane Library were searched up to May 2017. The following terms were used: (“coronary artery disease” or “coronary heart disease”) and (“X-ray repair cross complementing 1” or “XRCC1”). We also searched the references of the identified articles and relevant reviews.
Inclusion criteria
Case-control studies were considered to include in this meta-analysis. The case-control studies should have the genotype frequencies. Studies were excluded if they reported data overlapping with those of a larger study.
Extraction of data
Data were extracted as follow: first author, ethnicity, year, age, gender, the sample sizes, the polymorphisms.
Statistical analysis
Review Manager V.5.2 (The Cochrane Collaboration) was employed to analyse the pooled effects with Odds Ratio (OR) with 95% Confidence Interval (CI).. The genotype distribution was assessed by Hardy-Weinberg Equilibrium (HWE). Heterogeneity was assessed using the χ2-base Q test the I2 test. A P value less than 0.10 or an I2 more than 50% suggests significant heterogeneity. We used the fixed effect model for pooled analysis preferentially; if high heterogeneity was identified, we used the random effect model instead. We also did subgroup analyses according to race. Statistical significance was considered when a 2-tailed P value less than 0.05 was observed.
Results
Characteristics of the studies
Five studies (1715 cases and 1520 controls) were included in this study. Two case-control studies were done in Asians. Three case-control studies were done in Caucasians. All studies investigated rs25487 and two studies investigated rs1799782. Table 1 showed the main characteristics.
| Study | Year | Ethnicity | Age group | Gender | No. of case | No. of control | Polymorphisms |
|---|---|---|---|---|---|---|---|
| Bazo | 2011 | Caucasian | Adult | Mixed | 117 | 52 | rs25487, rs1799782 |
| Gokkusu | 2013 | Caucasian | Adult | Mixed | 197 | 135 | rs25487 |
| Narne | 2013 | Asian | Adult | Mixed | 160 | 121 | rs25487 |
| Yu | 2014 | Asian | Adult | Mixed | 1142 | 1106 | rs25487 |
| Hameed | 2016 | Caucasian | Adult | Mixed | 99 | 106 | rs25487, rs1799782 |
Table 1. Characteristics of the studies included in this meta-analysis.
Results of this study
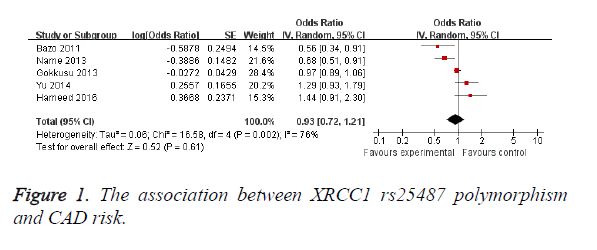
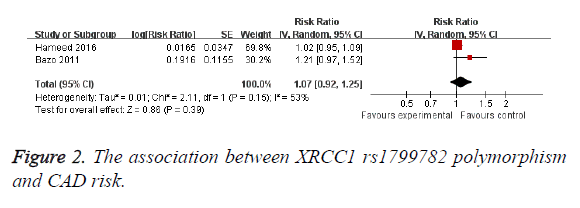
The results are shown in Table 2. No significant association was found between XRCC1 rs25487 polymorphism and CAD (OR=0.93; 95% CI, 0.72-1.21; P=0.61; Figure 1). When stratified by race, no significant association was found between XRCC1 rs25487 polymorphism and CAD risk in Caucasian (OR=0.93; 95% CI, 0.63-1.39; P=0.74; Table 2) and Asian (OR=0.93; 95% CI, 0.50-1.75; P=0.83; Table 2). The shape of the funnel plot did not show obvious asymmetry (data not shown). A significant association was also not found between XRCC1 rs1799782 polymorphism and CAD risk (OR=1.07; 95% CI, 0.92-1.25; P=0.39; Figure 2). In the Caucasian subgroup, no significant association was observed (OR=1.07; 95% CI, 0.92-1.25; P=0.39). Funnel plot also could not find obvious asymmetry.
| Polymorphism | No. of studies | Test of association | Heterogeneity | ||
|---|---|---|---|---|---|
| OR (95% CI) | P value | I2 (%) | P value | ||
| rs25487 | 5 | 0.93 (0.72-1.21) | 0.61 | 76 | 0.002 |
| Caucasian | 3 | 0.93 (0.63-1.39) | 0.74 | 74 | 0.02 |
| Asian | 2 | 0.93 (0.50-1.75) | 0.83 | 88 | 0.004 |
| rs1799782 | 2 | 1.07 (0.92-1.25) | 0.39 | 53 | 0.15 |
| Caucasian | 2 | 1.07 (0.92-1.25) | 0.39 | 53 | 0.15 |
Table 2. Result of this meta-analysis.
Discussion
No significant association was found in this study. When stratified by race, XRCC1 rs25487 polymorphism has not influence the CAD risk in Caucasian and Asian. Additionally, XRCC1 rs1799782 polymorphism has not influence the CAD risk in Caucasian. Our results were different from the previous two meta-analyses [14,15]. Both of them found positive results. However, they included wrong studies. Thus, the results from their studies were not reliable.
XRCC1 Arg399Gln polymorphism was associated with hepatitis-related hepatocellular carcinoma [16]. Jiang et al. suggested that pancreatic cancer risk was associated with XRCC1 rs25487 polymorphism in Asians [17]. Wang et al. suggested that lung cancer risk in Asian population was associated with XRCC1 399 polymorphism [18]. Yang et al. indicated that bladder cancer might be associated with XRCC1 rs25487 polymorphism [19]. Chen et al. concluded that laryngeal cancer was not associated with XRCC1 Arg399Gln polymorphism [20].
Some limitations should be addressed. First, more case-control studies are needed to assess the results in different races. Second, publication bias cannot be ruled out. Third, we did not perform other subgroup analyses due to limited data.
In conclusion, this meta-analysis suggested that XRCC1 gene rs25487, rs1799782 polymorphisms were not associated with CAD risk.
Disclosure of Conflict of Interest
The authors have declared that no competing interests exist.
References
- GBD 2015 DALYs, HALE Collaborators. Global, regional, and national disability-adjusted life-years (DALYs) for 315 diseases and injuries and healthy life expectancy (HALE), 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016; 388: 1603-1658.
- Frazier L, Johnson RL, Sparks E. Genomics and cardiovascular disease. J Nurs Scholarsh 2005; 37: 315-321.
- Ozaki K, Tanaka T. Molecular genetics of coronary artery disease. J Hum Genet 2015; 60: 1715-1721.
- Fan J, Wilson DM. Protein-protein interactions and posttranslational modifications in mammalian base excision repair. Free Radic Biol Med 2005; 38: 1121-1138.
- Forat-Yazdi M, Gholi-Nataj M, Neamatzadeh H, Nourbakhsh P, Shaker-Ardakani H. Association of XRCC1 Arg399Gln polymorphism with colorectal cancer risk: a HuGE meta analysis of 35 studies. Asian Pac J Cancer Prev 2015; 16: 3285-3291.
- Yan J, Wang X, Tao H, Deng Z, Yang W, Lin F. Meta-analysis of the relationship between XRCC1-Arg399Gln and Arg280His polymorphisms and the risk of prostate cancer. Sci Rep 2015; 5: 9905.
- Fan X, Xiu Q. Effect of X-ray repair cross complementing group 1 polymorphisms on the efficacy of platinum-based chemotherapy in patients with nonsmall cell lung cancer. J Cancer Res Ther 2015; 11: 571-574.
- Sanjari MA, Nazarzadeh M, Sanjari Moghaddam H, Bidel Z, Karamatinia A, Darvish H, Mosavi-Jarrahi A. XRCC1 gene polymorphisms and breast cancer risk: a systematic review and meta- analysis study. Asian Pac J Cancer Prev 2016; 17: 323-330.
- Bazo AP, Salvadori D, Salvadori RA, Sodre LP, da Silva GN, de Camargo EA, Ribeiro LR, Salvadori DM. DNA repair gene polymorphismis associated with the genetic basis of atherosclerotic coronary artery disease. Cardiovasc Pathol 2011; 20: 9-15.
- Gokkusu C, Cakmakoglu B, Dasdemir S, Tulubas F, Elitok A, Tamer S, Seckin S, Umman B. Association between genetic variants of DNA repair genes and coronary artery disease. Genet Test Mol Biomarkers 2013; 17: 307-313.
- Narne P, Ponnaluri KC, Singh S, Siraj M, Ishaq M. Arg399Gln polymorphism of X-ray repair cross-complementing group 1 gene is associated with angiographically documented coronary artery disease in South Indian type 2 diabetic patients. Genet Test Mol Biomarkers 2013; 17: 236-241.
- Yu X, Liu J, Zhu H, Xia Y, Gao L, Dong Y, Jia N, Shen W, Yang Y, Niu W. Synergistic association of DNA repair relevant gene polymorphisms with the risk of coronary artery disease in northeastern Han Chinese. Thromb Res 2014; 133: 229-234.
- Hameed H, Faryal M, Aslam MA, Akbar A, Saad AB, Pasha MB, Latif M, Rehan Sadiq Shaikh RR, Ali M, Iqbal F. Association of polymorphisms (rs 1799782, rs25489 and rs25487) in XRCC1 and (rs 13181) XPD genes with acute coronary artery syndrome in subjects from Multan, Pakistan. Pak J Pharm Sci 2016; 29: 869-876.
- Guo SJ, Zhou YT, Liu WY, Zuo QN, Li XH. The polymorphism of XRCC1 and coronary artery disease risk: a meta-analysis. Eur Rev Med Pharmacol Sci 2017; 21: 1559-1567.
- Ma WQ, Han XQ, Wang X, Wang Y, Zhu Y. Associations between XRCC1 gene polymorphisms and coronary artery disease: a meta-analysis. PLoS One 2016; 11: e0166961.
- Wu D, Jiang H, Gu Q, Zhang D, Li Z. Association between XRCC1 Arg399Gln polymorphism and hepatitis virus-related hepatocellular carcinoma risk in Asian population. Tumour Biol 2013; 34: 3265-3269.
- Jiang H, Wu D, Ma D, Lin G, Liang J, Jin J. Association between X-ray repair cross-complementation group 1 rs25487 polymorphism and pancreatic cancer risk. Tumour Biol 2013; 34: 3417-3421.
- Wang JY, Cai Y. X-ray repair cross-complementing group 1 codon 399 polymorphism and lung cancer risk: an updated meta-analysis. Tumour Biol 2014; 35: 411-418.
- Yang D, Liu C, Shi J, Wang N, Du X. Association of XRCC1 Arg399Gln polymorphism with bladder cancer susceptibility: a meta-analysis. Gene 2014; 534: 17-23.
- Chen W, Wang ZY, Xu FL, Wu KM, Zhang Y, Xu L, Wang QP. Association of XRCC1 genetic polymorphism (Arg399Gln) with laryngeal cancer: a meta-analysis based on 4,031 subjects. Tumour Biol 2014; 35: 1637-1640.