Research Article - Biomedical Research (2018) Volume 29, Issue 10
Validity of chromosomal aneuploidies testing during pregnancy: A comparison of karyotype, interphase-FISH and QF-PCR techniques
Hamid Galehdari1*, Mojgan Barati2, Mandana Mahmoudi2, Nahid Shahbazian2, Sara Masihi2, Mina Zamani3, Anise Poshtkuhian3, Marzieh Mohammadi Anaee3, Alireza Sedaghat3,4, Alihossein Saberi5, Mohammad Hamid6, Gholamreza Shariati3,5
1Thalassemia and Hemoglobinopathy Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
2Faculty of Medicine, Fertility, Infertility and Perinatology Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
3Narges Genetic Laboratory, Ahvaz, Iran
4Diabetes Research Center, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
5Department of Genetics, Faculty of Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
6Department of Molecular Medicine, Biotechnology Research Center, Pasteur Institute of Iran, Tehran, Iran
- *Corresponding Author:
- Hamid Galehdari
Thalassemia and Hemoglobinopathy Research Center
Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
Accepted on April 09, 2018
DOI: 10.4066/biomedicalresearch.29-17-3995
Visit for more related articles at Biomedical ResearchAbstract
Prenatal diagnosis of chromosomal abnormalities is an important challenge for pregnancy management has relied on conventional cytogenetic analysis of cultured amniotic fluid, chorionic villi, or fetal blood for a long time. New molecular methods included FISH and QF-PCR on uncultured amniotic fluid or chorionic villi have been applied recently for the rapid aneuploidies detection in chromosomes 13, 18, 21, X and Y. Both molecular methods are quicker than Karyotype as conventional cytogenetic technique. The advantages of QF-PCR include detection of trisomy, mosaicism and maternal cell contamination. This technique is more cost effective, less labor-intensive and more suitable for large sample numbers in comparison to FISH. FISH is a robust method and extensively validated. It detects mosaicism and trisomy. Herein we aimed to assess prenatal diagnosis of aneuploidies in chromosomes 13, 18, 21, X and Y. For this aim, amniotic fluid samples have collected for analysis of common chromosomal aneuploidies using Karyotype, FISH and QF-PCR methods. 69 samples have obtained by amniocentesis from 15th-18th week of pregnancies and subjected for comparative analysis by three techniques: FISH, QF, and Karyotype with the same accuracy concerning the detection of trisomy 13, 18, 21, and sex chromosomes X and Y. We conclude that molecular techniques are rapid-reliable methods for analysis of chromosome number in uncultured amniotic fluid. Nevertheless, QF-PCR and FISH are an alternative option, if timing is a limiting factor. This report in line with previous studies emphasizes the importance of molecular techniques for the prenatal identification of the numerical chromosomal abnormalities and ultimately management of pregnancies saving patients from anxiety.
Keywords
Prenatal testing, Aneuploidies, Molecular cytogenetics, Rapid diagnosis.
Introduction
Prenatal diagnosis of critical conditions especially chromosomal aneuploidies has been a concern through the decades. Accounting for more than 95% of live-born abnormalities in chromosomes, the most frequent aneuploidies prenatally diagnosed include Trisomy 21 (Down syndrome), Trisomy 18 (Edward syndrome), Trisomy 13 (Patau syndrome) and sex chromosome aneuploidies [1,2]. Common indications for such abnormalities include advanced maternal age (≥ 35 y), family history of genetic diseases, multiple abortions, abnormal results of ultrasound findings and maternal serum screenings [3]. The traditional gold standard technique for prenatal detection of aneuploidies is analysis of metaphase chromosomes, which is known as karyotyping of cultured amniotic fluid cells (amniocentesis) or chorionic villi cells with up to 99.5% accuracy [4,5]. In spite of practicability of karyotyping as a cytogenetic technique for prenatal diagnosis of chromosomal aberrations in laboratories, this traditional method is associated with considerable limitations such as time-consuming and laborious cell cultivation, low resolution of the cell-treated field, and limitation in detection of chromosome abnormalities which are less than 5 mb in length [5,6]. As a result, sophisticated substitutions like fluorescent in situ hybridization (FISH) and quantitative fluorescence polymerase chain reaction (QF-PCR) using uncultivated amniotic fluid came through investigations [7].
Interphase-FISH method accomplished on uncultured cells was developed in 1997 [8,9], which applies Interphase rather than metaphase cultured-cells in metaphase-FISH, is a rather privileged method for rapid and early prenatal aneuploidies screening with high sensitivity and accuracy [10,11]. However, interphase FISH has an implied disadvantage with regard to directly detection of whole chromosomes or chromosome regions, yet sufficient to distinguish between polyploid, monoploid, and diploid cells [12].
The two above mentioned methods (QF-PCR and FISH) culminate at about 24-48 h which is satisfying for the stressed patients waiting for the results [13].
We routinely perform karyotype in our center as gold standard for aneuploidy detection, which takes between 10-12 working days. This time would be seemingly very long for pregnant stressed women. Our aim was to introduce a rapid pre-test for most common trisomy 21, 18, and 13 along to the karyotype. For these reasons, the present study was designed for testing the validity of new molecular cytogenetic techniques like interphase-FISH and QF-PCR as chromosomal aberration screening tests in high-risk pregnancies indicated as advanced maternal age>35 y and/or positive maternal serum screen.
Materials and Methods
Sample preparation
Pregnant women had attended to the Perinatology department of Ahvaz Imam Khomeini Hospital. Women with low or intermediate-risk (by routine screening test) were excluded for this study. After informed consent, 69 amniotic fluid samples were collected from volunteer high-risk women (1st and 2nd trimester screening under cut-off) from 15th to 18th weeks of pregnancy in a period of 3 months. Obtained fetal cells were divided in two parts: one part was hold for culturing, and other part was directly used for FISH and QF-PCR. All cytogenetic and molecular tests were performed in an accredited genetic laboratory specified for prenatal screening and diagnosis.
Karyotype
Fetal cells obtained from amniocentesis have cultured in amnioMax media (Invitrogen Co.), harvested, fixed and synchronized in metaphase by colchicines. Chromosomes have stained and aligned pair wise by special software (Cytovision, USA) and analyzed finally.
FISH
FISH for common aneuploidy of chromosomes 21, 18, 13 and sex chromosomes X and Y was performed using Poseidon FISH Kit according to the manufacturer's instructions (Kreatech, Amsterdam, Netherlands).
Samples were centrifuged and incubated in hypotonic solution and subsequent fixative at a 3:1 methanol-to-acetic acid ratio for 30 min for FISH. The latter slides were air-dried and stored at -20°C until pre-treatment and hybridization for FISH. Four slides were prepared for each specimen. After slide preparation, 3-μl probe was hybridized in 37°C for 15 h; ultimately, slides were washed and signals observed using florescent microscope. Additionally, accuracy and positions of signals were analyzed by cytovision (USA) software.
Quantitative fluorescent PCR
QF-PCR is a PCR-based technique that consists of amplifying polymorphic markers located on the chromosomes of interest to determine the number of copies of those chromosomes present per cell. For this propose, fetal genomic DNA was extracted from 5 ml amniotic fluid with routine procedures. QF-PCR procedure was done according to the manufacturer's instructions. Optimizing of DNA concentration for QF-PCR was achieved by serial dilution of exact DNA amount from whole blood
Results
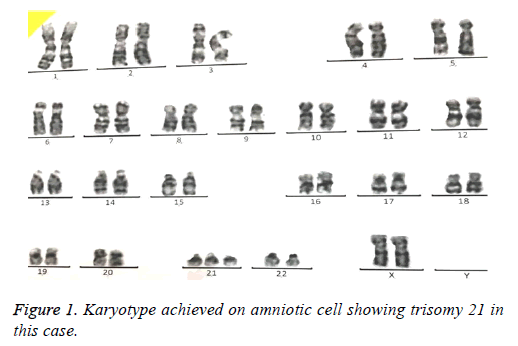
69 amniotic fluid samples have been collected for prenatal analysis of common and rare aneuploidies. In average, 15-20 ml amniotic fluid was taken from pregnant women. Karyotype was performed as gold standard for all samples were cultured 5-10 d depending on cell number. At least, 30 cells were analyzed by computational software (Figure 1). In case of nonoptimal spreads so that analysis by computer was not possible, an expert did manual analysis of chromosome banding.
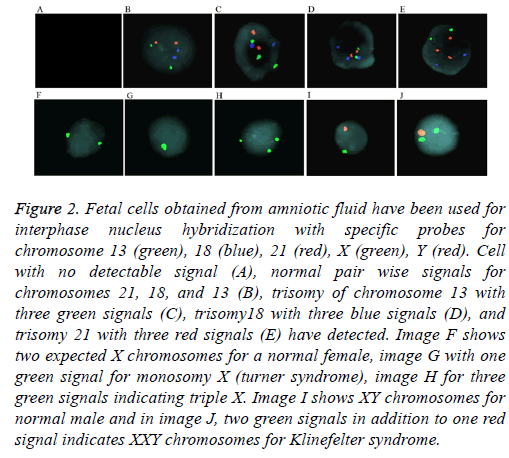
For aneuploidy detection using FISH, five specific probes 21, 18, 13, and sex chromosomes X and Y were used (Figure 2). Pre-hybridization and hybridization time of probes was constant for all samples. FISH signals were declared as true if detectable easily, in which we excluded noisy weak signals Only non-overlapping cells were scored. Criteria for Trisomy or monosomy are explained in Figure 2. The entire FISH assay took 2 working days. From 69 samples, 64 samples were with detectable signals (93%) and five samples were noninformative (7%) with noisy non-optimized signals. These 5 samples had normal karyotype and QF-PCR results. Standard error for entire samples and five probes (21, 18, 13, X, and Y) was estimated as 0.2%.
Figure 2: Fetal cells obtained from amniotic fluid have been used for interphase nucleus hybridization with specific probes for chromosome 13 (green), 18 (blue), 21 (red), X (green), Y (red). Cell with no detectable signal (A), normal pair wise signals for chromosomes 21, 18, and 13 (B), trisomy of chromosome 13 with three green signals (C), trisomy18 with three blue signals (D), and trisomy 21 with three red signals (E) have detected. Image F shows two expected X chromosomes for a normal female, image G with one green signal for monosomy X (turner syndrome), image H for three green signals indicating triple X. Image I shows XY chromosomes for normal male and in image J, two green signals in addition to one red signal indicates XXY chromosomes for Klinefelter syndrome.
The total nucleated cells in each ml of amniotic fluid vary between 103-106 cells [8]. Here, genomic DNA extraction of fetal cells from 5 ml amnion fluids as well as two small chorionic villi fronds was sufficient for reliable results in QFPCR method. From 69 samples, 65 samples were informative (95%), and further four samples were non-informative (5%). These four samples had normal karyotype and FISH results.
In summary, results from sixty samples were in concordance in all three methods compared to each other. In other nine cases, at least two of three methods confirmed each other. In such cases, results of third method failed, because of eventually low quality of probes or false calculating of probe concentration (FISH), or limiting DNA concentration (QF-PCR).
In conclusion, 8 samples were positive for trisomy 21, 5 for trisomy 18, and 3 cases for trisomy 13. Six samples were positive for Turner syndrome, four for Klein-felter syndrome, and two for triple X. All other samples showed normal number of chromosomes.
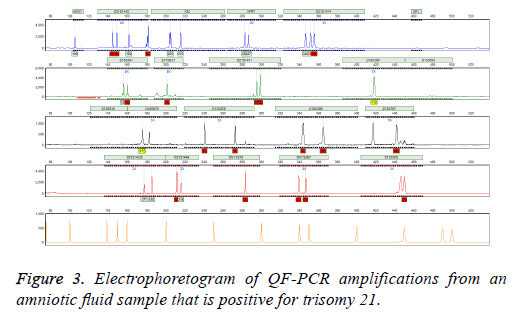
Follow-up Karyotype or array CGH confirmed the results of three mentioned methods after giving birth or after abortion therapy. In case of detected aneuploidy, we used aborted material for confirming tests. In case of normal diagnosis, we used neonatal blood after birth for karyotype. We observed no false positive or false negative results in entire specimens. In addition, our results were finally validated by clinical diagnosis after delivery (Figure 3).
Discussion
Prenatal diagnosis of chromosomal abnormalities is one of the noticeable challenges in perinatology. The most frequent aneuploidies prenatally diagnosed include trisomy 13, 18, 21 and sex chromosome aneuploidies [1,2]. Rapid and accurate assessment of chromosomal abnormalities was necessary to manage pregnancy. This involves some conventional cytogenetic (karyotype) and new molecular cytogenetic techniques (FISH and QF-PCR) for this end.
Standard cytogenetic method known as karyotype is associated with some disadvantages including technical difficulties, long required time to obtain the test results because of the sufficient cell number requirement and interpretation of the resulting banded chromosome preparations, and false positive result. The delay can cause women under anxiety. On the other hand, the advantage of molecular cytogenetic techniques included FISH and/or QF-PCR is quick reporting within 2 d and earlier relief of anxiety. Also the accuracy and reliability of these molecular techniques have been demonstrated and misdiagnosis by either is small [13,14]. These techniques have inherent limitation as they can detect only structural chromosome abnormalities for which the primers and probes are designed. These methods are used mainly as a prelude to karyotyping. However, there are challenges about the potential advantage for introducing a stand-alone test for prenatal chromosome aneuploidies diagnosis for low risk pregnancies [14,15].
To the conventional karyotype as a gold standard technique for aneuploidies, we compared here two further methods FISH and QF-PCR regarding their time consuming, senstivity, specifity, and detecting capacity. However, our general aim was here to accelerate the detection of common aneuploidy in high-risk pregnancies as nuchal translucency greater than 3.5, advanced maternal age (>35 y), and/or positive serum screening, and not to replace karyotype with other techniques.
Regarding QF-PCR, genomic DNA from few fetal cells was sufficient to perform it, which was advantageous in cases with deficiency of amniotic fluid (oligohydramnios). We estimated that less than one ng DNA is sufficient to get robust results. Further benefit of QF-PCR was its fast execution (2 h DNA extraction, 3 h PCR, and 5 h analysis of results). Under optimized conditions, QF-PCR is able to detect other abnormalities than common and rare trisomies (depending on its STR primer capacity), which was not to be discussed here. QF-PCR has the advantage of being less expensive, less laborintensive and provides simultaneously processing of larger number of samples. In addition, QF-PCR can detect mosaicism and maternal cell contamination when trisomy is present in more than 10% of in vitro cultured cells [16,17]. Recent report suggest that by using QF-PCR as a stand-alone test, the chances of non-diagnosing the commonest, and the only chromosome anomalies which do increase in frequency with maternal age, are approximately one in 150 abnormal karyotypes, or one in 10-30000 samples, based on the age distribution. These error rates might be deemed acceptable [18].
Interphase-FISH technique is nowadays applied routinely for rapid testing of uncultured cells to identify chromosomes aneuploidies. The accuracy of FISH is depending largely on specific probes, and a wide range of probe types can be used from whole genomes to small cloned probes [19]; Chase et al. reported that incorrect choice of locus specific probe might lead to false positive or negative results [20]. However, the high sensitivity and specificity of FISH probes and the performance speed of technique have made it a robust cytogenetic test lead to significant advances in both the research and clinical applications [21]. Many more recent literature reports have indicated a much higher informativeness. They also show that the risk of either over or under-diagnosis of aneuploidy for the target chromosomes is small by interphase FISH, using the ‘gold standard’ of karyotyping as a comparison [22].
It should be noted that although molecular methods that allow rapid and reliable prenatal diagnosis of targeted fetal chromosome abnormalities, for a small proportion of samples do not provide an unequivocal diagnostic. Co-application of karyotyping with one of these tests is recommended for obtaining result that is more accurate [23]. Therefore and because of worse consequences in case of false negative results, we suggest performing karyotype for any case, with application of second test as pre-test (FISH or QF-PCR) for fast preliminary results to calm down stressed high-risk women.References
- Whiteman DAH, Klinger K. Efficiency of rapid in situ hybridization methods for prenatal diagnosis of chromosome abnormalities causing birth defects. Am J Hum Genet 1991; 49: A1279.
- Divane A. Rapid prenatal diagnosis of aneuploidy from uncultured amniotic fluid cells using five-colour fluorescence in situ hybridization. Prenat Diagn 1994; 14: 1061-1069.
- Binns V, Hsu N. Prenatal diagnosis. Encyclopedia Life Sci 2002.
- Lim AS. Rapid aneuploidy screening with fluorescence in-situ hybridisation: is it a sufficiently robust stand-alone test for prenatal diagnosis? Hong Kong Med J 2010; 16: 427-433.
- Leung WC. Rapid aneuploidy testing, traditional karyotyping, or both, in prenatal diagnosis. Hong Kong J Gynaecol Obstetr Midwifery 2005; 5: 33-39.
- Hurd D. Delving deep into the genetic basis of disease. Am Biotechnol Laboratory 2010.
- Nemescu D. Fetal aneuploidy diagnosis through rapid fluorescence in situ hybridization (FISH) on uncultured amniocytes. Revista Românăde Medicinăde Laborator 2011; 19: 161-167.
- Milunsky A. Genetic disorders and the fetus: diagnosis, prevention and treatment. Baltimore: The Johns Hopkins University Press 1998:128-149.
- Gersh M. Development of diagnostic tools for the analysis of 5p deletions using interphase FISH. Cytogenetics Cell Genet 1997; 77: 246-251.
- Xu J, Chen Z. Advances in molecular cytogenetics for the evaluation of mental retardation. Am J Med Genet 2003; 117: 15-24.
- Désilets VA. Use of a DNA method, QF-PCR, in the prenatal diagnosis of fetal aneuploidies. Joint Sogc-Ccmg Clin Pract Guideline 2011; 33: 955-960.
- David AE, Maik S, Rupa DS. Advantages and limitations of using fluorescence in situ hybridization for the detection of aneuploidy in interphase human cells: Mutation Res 1995; 348: 153-162.
- Riegel M. Human molecular cytogenetics: From cells to nucleotides. Genet Mol Biol 2014; 37: 194-209.
- Leung WC, Lao TT. Rapid aneuploidy testing, traditional karyotyping, or both? Lancet 2005; 366: 97-98.
- Hulten MA, Dhanjal S, Pertl B. Rapid and simple prenatal diagnosis of common chromosome disorders: advantages and disadvantages of the molecular methods FISH and QF-PCR. Reproduction 2003; 126: 279-297.
- Halder A. Fluorescence in situ hybridization (FISH) using non-commercial probes in the diagnosis of clinically suspected micro-deletion syndromes. Indian J Med Res 2013; 138: 135-142.
- Shaffer LG, Bui TH. Molecular cytogenetic and rapid aneuploidy detection methods in prenatal diagnosis. Am J Med Genet 2007; 145: 87-98.
- Ogilvie CM. Prenatal diagnosis for chromosome abnormalities: past, present and future. Pathol Biol 2003; 51: 156-160.
- Mann K, Ogilvie CM. QF-PCR: application, overview and review of the literature. Prenat Diagn 2012; 32: 309-314.
- Umberto N, Faustina L, Federica N, Cristina C, Hung B. The introduction of QF-PCR in prenatal diagnosis of fetal aneuploidies: time for reconsideration. Human Reprod Update 2004; 10: 541-548.
- Ryan B. Applications of fluorescence in situ hybridization (FISH) in detecting genetic aberrations of medical significance. Biosci Horizons 2010; 3: 85-95.
- Chase A, Grand F, Zhang JG. Factors influencing the false positive and negative rates of BCR-ABL fluorescence in situ hybridization. Genes Chromosomes Cancer 1997; 18: 246-253.
- Caine A. Prenatal detection of Down's syndrome by rapid aneuploidy testing for chromosomes 13, 18, and 21 by FISH or PCR without a full karyotype: a cytogenetic risk assessment. Lancet 366: 123-128.