Short Communication - Biomedical Research (2018) Volume 29, Issue 8
Validation of anti HCV-hepatitis c identification in blood donors using PCR: case study at Tuban East Java district, Indonesia
Supiana Dian Nurtjahyani1*, Retno Handajani2, Mochammad Amin3, Dewi Hidayati4 and Nurul Jadid4
1Faculty of Teacher Training and Education, Universitas PGRI Ronggolawe, Tuban, Indonesia
2Faculty of Medicine, University of Airlangga, Surabaya, Indonesia
3Institute of Tropical Disease, University of Airlangga, Surabaya, Indonesia
4Department of Biology, Institute Teknologi Sepuluh Nopember (ITS), Surabaya, Indonesia
- *Corresponding Author:
- Supiana Dian Nurtjahyani
Faculty of Teacher Training and Education
Universitas PGRI Ronggolawe, Indonesia
Accepted date: February 20, 2018
Keywords
Blood donors, ELISA, PCR, Anti HCV virus, Tuban
Introduction
Virus infection caused by hepatitis C virus (HCV) is still an issue on global health. Hepatitis C virus is an RNA virus from hepacivirus family, single stranded and have similarity with flavivirus [1,2]. Since the hepatitis C virus can be transmitted from blood transfusion (90% from cases), the evaluation of HCV in the blood among donors is crucially important [2,3]. ELISA is common methods for HCV detection, however inaccurate result possibly occurred during the window period, hence the validation by re-testing this method using molecular test using Polymerase Chain Reaction (PCR) is still required [4,5].
This research would be useful for the local government as scientific recommendation to include the PCR validation test into their Standard Operating Procedure (SOP) of screening blood donors. This case report was conducted in Tuban as a representative region in East Java, Indonesia.
Material and Methods
Sample collection
Blood samples were collected from 100 donors in Tuban region according to standard procedure from the Indonesian Red Cross Society.
Hepatitis C identification
Serum from 100 blood donors was then subjected to Anti-HCV identification using Enzyme-Linked Immunosorbent Assay (ELISA), following the instructions (HCV Combo, Biorad, Germany).
Furthermore, the serum samples with negative anti-HCV were prepared for retesting using PCR analysis. Prior to PCR analysis, total RNA-virus in the serum was extracted using RNeasy mini kit -QIAGEN protocols.
The extracted RNA virus was then converted into complementary DNA (cDNA) refers to ReverTra Ace® qPCR Master mix (Toyobo) as previously described by Jadid et al. [6] for further PCR amplification. The reaction was performed within two rounds of PCR. Detail primers in Table 1 [7].
| 166 | TGGGGATCCCGTATGATACCCGCTGCTTTGA | 8230-8260 | + | Outer |
| 167R | GGCGGAATTCCTGGTCATAGCCTCCGTGAA | 8601-8630 | - | |
| HC23 | TTTGACTCAACCGTCACTGA | 8256-8275 | + | Inner |
| HC24 | CTCAGGCTCGCCGCATCCTC | 8577-8596 | - | |
| HC26 | CTCAGGTTCCGCTCGTCCTC | 8577-8596 | - | |
| HC15 | ACTGTCACTGAACAGGACAT | 8265-8284 | + | Inner |
| HC16 | GCTCTATCCTCATCGACGCC | 8568-8587 | - | |
| HC28 | CACGAGCATGGTGCAGTCCCGGAGC | 8507-8531 | - | |
| HC32 | AGGTAGCACGTCAGCGTGTTTCC | 8454-8476 | - | |
| HC34 | TAGCACGTCATGGTGTTTCCCAT | 8451-8473 | - |
Table 1. Details of the primers for HCV amplification.
PCR used “GeneAmp PCR System 2400 (Perkin Elmer)” Every PCR conducted using 40 cycles. Before the cycle hot start conducted with temperature of 94ºC for 5 min. Condition of the PCR cycles describe below: denaturation at 94ºC for 60 s, annealing temperature for 60 s and elongation at 72ºC for 75 s.
Amplification result visualization
PCR results were visualized by gel electrophoresis which consist of TBE buffer 0.5X, 2% of agarose gel and supplemented with ethidium bromide. The 1000 bp DNA ladder HAE III Digest was used as DNA marker.
Data analysis
Data population of HCV infected status of blood samples by ELISA methods were qualitatively grouped for further investigation using PCR methods. Positive HCV determination based on PCR were analysed descriptively refer to visual DNA-virus bands.
Result and Discussions
The 100 blood samples were obtained from 85 male and 15 female donors. Blood sample HCV detection (positive and negative status) by ELISA was exhibited in following Table 2. The ELISA analysis indicated that all donors (100%) were negative HCV. These negative HCV were then administered with PCR analysis.
| Gender | Anti HCV | PCR | ||
|---|---|---|---|---|
| Ratio Negative/Positive | Percentage of negative HCV | Ratio Negative/Positive | Percentage of negative HCV | |
| Male | 85/100 | 85% | 5/85 | 5.8% |
| Female | 15/100 | 15% | 1/15 | 6.7% |
| Total | 100 | 100 % | 6/100 | 6% |
Table 2. Result of anti-HCV analysis using ELISA.
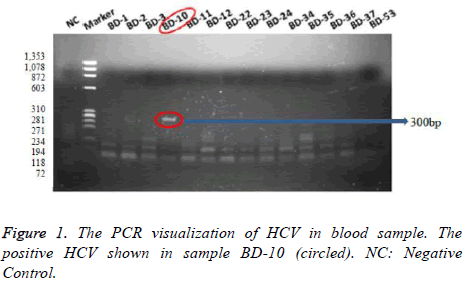
The PCR results showed that 5 samples (5.8%) among 85 negative samples of male 1 sample (6.7%) among 15 negative samples of male were detected positively infected by HCV. The PCR visualizations of HCV in blood sample were represented Figure 1, which positive one was shown in sample BD-10 (300 bp).
The positive HCV that resulted by PCR analysis mainly depends on the choice of selected primer. In this study, we used primer which was designed based on the sequences of nonstructural protein 5B (NS5B) of hepatitis C virus (HCV) [8]. NS5B have a polymerase activity which responsible for viral genome replication [9]. Moreover, due to its capacity, the NS5B has been developed for antivirus therapy. Some studies reported NS5B region can be used to determine genotype and subtype of hepatitis C virus as well as an effective learning tool for molecular HCV epidemiology [10,11]. This case study demonstrated that 6% detection by ELISA were inaccurate. Since the HCV transmission is mediated by blood, this 6% value considerably crucial and highly risk. Accordingly, the validation using PCR for screening blood donors in Tuban region is necessarily required.
Acknowledgement
This study was financially supported by Ministry of Research, Technology, and Higher Education of Indonesia. The authors would like to thank to PMI Tuban blood donors and the Center of Blood Transfusion-Tuban for providing sample collection and Institute of Tropical Disease (ITD)-Universitas Airlangga for facilitating the laboratory experiments required for this study.
Conclusion
According to HCV analysis using ELISA toward 100 blood donors sample, there was resulted 100% negative HCV. After retested by PCR analysis, there were found 6 blood samples (6%) positively infected HCV. This study encourages who one concerned to propose that PCR-based validation for screening blood donors is necessarily required.
References
- Lavanchy D. Evolving epidemiology of hepatitis C virus. Clin Mcrobiol Infect 2011; 17: 107.
- Friedman LS. Liver, biliary tract and pancreas disorders. Current Medical Diagnosis and Treatment 52nd Edn.) New York 2013.
- Firdaus R, Saha K, Biswas A, Sadhukhan PC. Current molecular methods for the detection of hepatitis C virus in high risk group population: A systemic review. World J Virol 2015; 4: 25-32.
- Vrielink H, Reesink HL, Zaaijer HL, Scholten E, Kremer LC, Cuypers HT. Lookback of anti-HCV ELISA positive, HCV-RNA PCR-negative donors and recipients of their blood products. Vox Sang 1997; 72: 67-70.
- Marwaha N, Sachdev S. Current testing strategies for hepatitis C virus infection in blood donors and the way forward. World J Gastroenterol 2014; 20: 2948-2954.
- Jadid N, Mardika RK, Nurhidayati T, Irawan MI. Reverse transcription-PCR analysis of geranylgeranyl diphosphate synthase (JcGGPPS) in Jatropha curcas L. and in silico analysis of Casbene Synthase (JcCS) among Euphorbiaceae. AIP Conference Proc 2016; 1744: 1.
- Juniastuti UT, Nasronudin AL, Amin M. High rate of seronegative HCV infection in HIV positive patients. Biomed Rep 2014; 2: 79-84.
- Tong YQ, Liu B, Liu H, Zheng HY, Gu J, Liu H, Song EJ, Song C, Li Y. Accurate genotyping of hepatitis C virus through nucleotide sequencing and identification of new HCV subtypes in China population. Clin Microbiol Infect 2015; 21: 874.e9-874.e21.
- Zhong W, Ferrari E, Lesburg CA, Maag D, Ghosh SK, Cameron CE, Lau JY, Hong Z. Template/primer requirements and single nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase J Virol 2000; 74: 9134-9143.
- Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N. Consensus proposals for a unified system of nomenclature of Hepatitis C Virus genotypes. Hepatology 2005; 42: 962.
- Murphy DG, Willems B, Deschenes M, Hilzenrat N, Mousseau R, Sabbah S. Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 59 untranslated region sequences. J Clin Microbiol 2007; 45: 1102-1112.
DOI: 10.4066/biomedicalresearch.29-17-2693
Visit for more related articles at Biomedical ResearchAbstract
The validation of virus-free blood donors is often neglected possibly related to the high cost process analyzing. While, validity of the medical status of hepatitis C virus (HCV) is crucial for blood donors quality. ELISA is common method for HCV detection, however inaccurate result possibly occurred during the window period. Hence, the validation of this method using molecular test is still required. The samples were collected from 100 blood donors consist of 85 males (85%) and 15 females (15%) from Tuban region, East Java Indonesia. The ELISA kit test resulted 100% of negative HCV. Surprisingly, after validated using PCR there were found 6 blood samples (6%) positively infected HCV. This study encourages who one concerned to propose that PCR-based validation for screening blood donors is necessarily required.
Keywords
Blood donors, ELISA, PCR, Anti HCV virus, Tuban
Introduction
Virus infection caused by hepatitis C virus (HCV) is still an issue on global health. Hepatitis C virus is an RNA virus from hepacivirus family, single stranded and have similarity with flavivirus [1,2]. Since the hepatitis C virus can be transmitted from blood transfusion (90% from cases), the evaluation of HCV in the blood among donors is crucially important [2,3]. ELISA is common methods for HCV detection, however inaccurate result possibly occurred during the window period, hence the validation by re-testing this method using molecular test using Polymerase Chain Reaction (PCR) is still required [4,5].
This research would be useful for the local government as scientific recommendation to include the PCR validation test into their Standard Operating Procedure (SOP) of screening blood donors. This case report was conducted in Tuban as a representative region in East Java, Indonesia.
Material and Methods
Sample collection
Blood samples were collected from 100 donors in Tuban region according to standard procedure from the Indonesian Red Cross Society.
Hepatitis C identification
Serum from 100 blood donors was then subjected to Anti-HCV identification using Enzyme-Linked Immunosorbent Assay (ELISA), following the instructions (HCV Combo, Biorad, Germany).
Furthermore, the serum samples with negative anti-HCV were prepared for retesting using PCR analysis. Prior to PCR analysis, total RNA-virus in the serum was extracted using RNeasy mini kit -QIAGEN protocols.
The extracted RNA virus was then converted into complementary DNA (cDNA) refers to ReverTra Ace® qPCR Master mix (Toyobo) as previously described by Jadid et al. [6] for further PCR amplification. The reaction was performed within two rounds of PCR. Detail primers in Table 1 [7].
| 166 | TGGGGATCCCGTATGATACCCGCTGCTTTGA | 8230-8260 | + | Outer |
| 167R | GGCGGAATTCCTGGTCATAGCCTCCGTGAA | 8601-8630 | - | |
| HC23 | TTTGACTCAACCGTCACTGA | 8256-8275 | + | Inner |
| HC24 | CTCAGGCTCGCCGCATCCTC | 8577-8596 | - | |
| HC26 | CTCAGGTTCCGCTCGTCCTC | 8577-8596 | - | |
| HC15 | ACTGTCACTGAACAGGACAT | 8265-8284 | + | Inner |
| HC16 | GCTCTATCCTCATCGACGCC | 8568-8587 | - | |
| HC28 | CACGAGCATGGTGCAGTCCCGGAGC | 8507-8531 | - | |
| HC32 | AGGTAGCACGTCAGCGTGTTTCC | 8454-8476 | - | |
| HC34 | TAGCACGTCATGGTGTTTCCCAT | 8451-8473 | - |
Table 1. Details of the primers for HCV amplification.
PCR used “GeneAmp PCR System 2400 (Perkin Elmer)” Every PCR conducted using 40 cycles. Before the cycle hot start conducted with temperature of 94ºC for 5 min. Condition of the PCR cycles describe below: denaturation at 94ºC for 60 s, annealing temperature for 60 s and elongation at 72ºC for 75 s.
Amplification result visualization
PCR results were visualized by gel electrophoresis which consist of TBE buffer 0.5X, 2% of agarose gel and supplemented with ethidium bromide. The 1000 bp DNA ladder HAE III Digest was used as DNA marker.
Data analysis
Data population of HCV infected status of blood samples by ELISA methods were qualitatively grouped for further investigation using PCR methods. Positive HCV determination based on PCR were analysed descriptively refer to visual DNA-virus bands.
Result and Discussions
The 100 blood samples were obtained from 85 male and 15 female donors. Blood sample HCV detection (positive and negative status) by ELISA was exhibited in following Table 2. The ELISA analysis indicated that all donors (100%) were negative HCV. These negative HCV were then administered with PCR analysis.
| Gender | Anti HCV | PCR | ||
|---|---|---|---|---|
| Ratio Negative/Positive | Percentage of negative HCV | Ratio Negative/Positive | Percentage of negative HCV | |
| Male | 85/100 | 85% | 5/85 | 5.8% |
| Female | 15/100 | 15% | 1/15 | 6.7% |
| Total | 100 | 100 % | 6/100 | 6% |
Table 2. Result of anti-HCV analysis using ELISA.
The PCR results showed that 5 samples (5.8%) among 85 negative samples of male 1 sample (6.7%) among 15 negative samples of male were detected positively infected by HCV. The PCR visualizations of HCV in blood sample were represented Figure 1, which positive one was shown in sample BD-10 (300 bp).
The positive HCV that resulted by PCR analysis mainly depends on the choice of selected primer. In this study, we used primer which was designed based on the sequences of nonstructural protein 5B (NS5B) of hepatitis C virus (HCV) [8]. NS5B have a polymerase activity which responsible for viral genome replication [9]. Moreover, due to its capacity, the NS5B has been developed for antivirus therapy. Some studies reported NS5B region can be used to determine genotype and subtype of hepatitis C virus as well as an effective learning tool for molecular HCV epidemiology [10,11]. This case study demonstrated that 6% detection by ELISA were inaccurate. Since the HCV transmission is mediated by blood, this 6% value considerably crucial and highly risk. Accordingly, the validation using PCR for screening blood donors in Tuban region is necessarily required.
Acknowledgement
This study was financially supported by Ministry of Research, Technology, and Higher Education of Indonesia. The authors would like to thank to PMI Tuban blood donors and the Center of Blood Transfusion-Tuban for providing sample collection and Institute of Tropical Disease (ITD)-Universitas Airlangga for facilitating the laboratory experiments required for this study.
Conclusion
According to HCV analysis using ELISA toward 100 blood donors sample, there was resulted 100% negative HCV. After retested by PCR analysis, there were found 6 blood samples (6%) positively infected HCV. This study encourages who one concerned to propose that PCR-based validation for screening blood donors is necessarily required.
References
- Lavanchy D. Evolving epidemiology of hepatitis C virus. Clin Mcrobiol Infect 2011; 17: 107.
- Friedman LS. Liver, biliary tract and pancreas disorders. Current Medical Diagnosis and Treatment 52nd Edn.) New York 2013.
- Firdaus R, Saha K, Biswas A, Sadhukhan PC. Current molecular methods for the detection of hepatitis C virus in high risk group population: A systemic review. World J Virol 2015; 4: 25-32.
- Vrielink H, Reesink HL, Zaaijer HL, Scholten E, Kremer LC, Cuypers HT. Lookback of anti-HCV ELISA positive, HCV-RNA PCR-negative donors and recipients of their blood products. Vox Sang 1997; 72: 67-70.
- Marwaha N, Sachdev S. Current testing strategies for hepatitis C virus infection in blood donors and the way forward. World J Gastroenterol 2014; 20: 2948-2954.
- Jadid N, Mardika RK, Nurhidayati T, Irawan MI. Reverse transcription-PCR analysis of geranylgeranyl diphosphate synthase (JcGGPPS) in Jatropha curcas L. and in silico analysis of Casbene Synthase (JcCS) among Euphorbiaceae. AIP Conference Proc 2016; 1744: 1.
- Juniastuti UT, Nasronudin AL, Amin M. High rate of seronegative HCV infection in HIV positive patients. Biomed Rep 2014; 2: 79-84.
- Tong YQ, Liu B, Liu H, Zheng HY, Gu J, Liu H, Song EJ, Song C, Li Y. Accurate genotyping of hepatitis C virus through nucleotide sequencing and identification of new HCV subtypes in China population. Clin Microbiol Infect 2015; 21: 874.e9-874.e21.
- Zhong W, Ferrari E, Lesburg CA, Maag D, Ghosh SK, Cameron CE, Lau JY, Hong Z. Template/primer requirements and single nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase J Virol 2000; 74: 9134-9143.
- Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N. Consensus proposals for a unified system of nomenclature of Hepatitis C Virus genotypes. Hepatology 2005; 42: 962.
- Murphy DG, Willems B, Deschenes M, Hilzenrat N, Mousseau R, Sabbah S. Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 59 untranslated region sequences. J Clin Microbiol 2007; 45: 1102-1112.