Research Article - Biomedical Research (2017) Volume 28, Issue 15
Up-regulation of microRNA-375 inhibits the proliferation of gastric cancer cell through targeting AEG-1
Zhihui Cai and Shiyun Tan*
Department of Gastroenterology, Renmin Hospital of Wuhan University, Wuhan, Hubei Province, P.R. China
- *Corresponding Author:
- Shiyun Tan
Department of Gastroenterology
Renmin Hospital of Wuhan University, P.R. China
Accepted on May 10, 2017
Abstract
MicroRNAs (miRs) were a series of non-coding RNAs, which could suppress the transcription of mRNAs; the deregulations of miR could regulate the cancer cell proliferation and apoptosis. In the present study we try to investigate the biological effect of miR-375 on the gastric cancer cells. RT-PCR result showed that miR-375 expression was significantly decreased in gastric cancer cells compared with normal gastric cells and microRNA mimic transfection assay was used to investigate the biological effect of miR-375 in the gastric cancer cells. MTT results demonstrated that the induction of miR-375 significantly inhibited the gastric cancer cell proliferation and FACS study indicated the cell proliferation inhibition was attributed to the induction of gastric cancer cell apoptosis. Astrocyte elevated gene-1 (AEG-1) was an important oncogene in multiple aspects of cancer development and progression, our further studies indicated that the transfection of miR-375 mimic significantly decreased the expression of AEG-1 gene both in mRNA level and protein level, which suggest miR-375 regulate the gastric cancer cell through the regulation of AEG-1 gene. In conclusion, our findings suggested that miR-375 targeted AEG-1 gene in gastric cancer cell and regulated the gastric cancer cell growth in vitro, which highlight the therapeutic potential of miR-375 in gastric cancer clinical treatment.
Keywords
MicroRNA-375, Gastric cancer cell, Proliferation, Apoptosis, AEG-1.
Abbreviations
miR: MicroRNA; miR-375: MicroRNA-375; UTR: Untranslated Region; NGEC: Normal Human Gastric Epithelial Cells; PI: Propidium Iodide; AEG-1: Astrocyte Elevated Gene-1; mg: Milligram; mm: Micrometer; mol/L: Molar; %: Percent; kg: Kilogram; h: Hours; min: Minutes; ml: Milliliter.
Introduction
Gastric cancer was the one of the most common type of cancers, it could cause about 800,000 patients die every year. In general, the patients with advanced gastric cancer had a 5 y survival rate equal to 15% [1]. The clinical outcome of gastric cancer had gradually improved nowadays, but the prognosis is still unsatisfied. This is because the mechanism of the gastric cancer development was still poorly understood despite of a few oncogenes and tumor suppressor genes have been reported in gastric cancers [2,3]. Therefore further research of effective treatments for gastric cancer is required.
MicroRNAs (miRs) were a series of non-coding RNAs, which could suppress the transcription of mRNAs by partial complementary binding to 3' Untranslated Regions (UTRs), they were formed by about 21-24 nucleotides [4,5]. Recently studies showed that miR deregulations could regulate the cell proliferation and apoptosis [6,7]. Studies also reported that 43 miRs were up-regulated and 37 miRs were down-regulated in gastric cancer [8], which indicating the dysregulations of miRs were associated with the progression and prognosis of gastric cancer. A few studies indicated that miR-375 expression was down-regulated in many human cancers [9-11]; however the biological role of miR-375 in gastric cancer is still not be understood. In the present study we try to investigate the expression and mechanisms of miR-375 in gastric cancer, which could benefit to the treatment of gastric cancer.
Materials and Methods
Cell culture
SGC-7901, BGC-823 and MKN-28 cell lines were purchased from ATCC. Normal Human Gastric Epithelial Cells (NGEC) were established from gastric biopsy specimens obtained from upper gastrointestinal endoscopy as described previously [12]. The NGEC and the gastric cancer cell lines were cultured in DMEM (Invitrogen) medium supplemented with 10% fetal bovine serum (Invitrogen), 100 U/ml penicillin (Invitrogen) and 100 mg/ml streptomycin (Invitrogen).
Transfection assay
SGC-7901 cells were cultured overnight, then transfected the miR-375 mimic or control mimic into the cells (10 nM final concentration) by using Lipofectamine 3000, the control mimic was used as the control. Both miR-375 mimic and control mimics were purchased from Thermo Fisher.
Cell viability assay
MiR-375 mimic, control mimic transfected SGC-7901 cells and mock cells were cultured in 96-well plate (3500 cell/well); the MTT assay was performed to test the cell viability every 24 h. First MTT reagent was added in to the well at 1:10, and then incubated the cells for additional 4 h. The culture medium was removed and 50 μL DMSO was added. Absorbance was measured at 570 nm, with 655 nm as the reference wavelength. The absorbance was expressed as the cell viability.
MiR-375 quantification
Total RNA for miR-375 quantification was extracted from miR-375 mimic or control mimic transfected SGC-7901 cells using TRIzol reagent (Invitrogen) and reverse transcribed into cDNA with TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). The expression of miR-375 was assayed using the TaqMan MicroRNA Assays (Applied Biosystems, Foster City, CA, USA) specific for hsa-miR-375, and followed by real-time PCR using TaqMan Gene Expression Master Mix (Applied Biosystems). RUN6B RNA was used as a control. Expression of miR-375 with relative to U6 was determined using the 2-ΔΔCt method.
Cell apoptosis assay
MiR-375 mimic or control mimic transfected SGC-7901 cells were seeded into 6-well plates and cultured for 48 h. Then harvested the cells and stained the cells with Annexin V and Propidium Iodide (PI). Cell apoptosis was evaluated by using a flow cytometry equipped with CellQuest software (BD Biosciences).
qRT-PCR for astrocyte elevated gene-1 (AEG-1)
Total RNA was extracted from SGC-7901 cells and reverse transcribed into cDNA with the Reverse Transcriptase MMLV (Takara). qRT-PCR reactions were performed using the following parameters: 95°C for 10 min, followed by 40 cycles of 15 s at 95°C, and 30 s at 60°C. The primers used were showed as followed. AEG-1: 5’- GTGGTTCAACAGTATTGCGTTGTC-3’, 5’- AACATACTGCCATGTTCCTGAAAC-3’. GAPDH: 5’- GACTCATGACCACAGTCCATGC-3’, 5’- AGAGGCAGGGATGATGTTCTG-3’. GAPDH was used as the control.
Western blot assay
Protein was extracted from SGC-7901 cells, separated the protein and transferred the separated protein on to a PVDF membrane. Blocked the transferred PVDF membrane with block buffer for 1 h and incubated the membrane with primary antibody AEG-1 (Abcam) or GAPDH (Beyotime Biotechnology) for 2 h. Washing the PVDF membrane with TBST for 3 times and incubated the membranes with second antibody for 1 h. Washing the PVDF membrane with TBST for 3 times and the proteins were examined by using ECL chemiluminescence and exposed to X-ray film.
Statistical analysis
The statistical analysis was performed by using SPSS11.0 software. Data were showed as mean ± SD. Each experiment was performed in 3 independent studies. *P<0.05 indicated statistically significant.
Results
MiR375 was down-regulated in gastric cancer cells
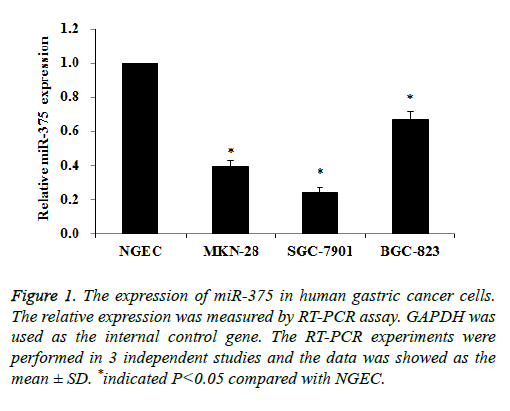
The expression of miR-375 in gastric cancer cells was measured by qRT-PCR assay, NGEC cells was used as the normal control. As indicated in Figure 1 the expression of miR-375 in gastric cancer cells was significantly decreased compared to NGEC cells. Especially the relative expression of miR-375 in SGC-7901 cells was 0.24 compared with NGEC cells (Figure 1). Therefore we suppose that the dysregulation of miR-375 was associated with the biological function of gastric cancer cells, and we chose SGC-7901 cells to investigate the hypothesis.
Figure 1: The expression of miR-375 in human gastric cancer cells. The relative expression was measured by RT-PCR assay. GAPDH was used as the internal control gene. The RT-PCR experiments were performed in 3 independent studies and the data was showed as the mean ± SD. *indicated P<0.05 compared with NGEC.
Up-regulation of miR-375 decreases gastric cancer cells proliferation
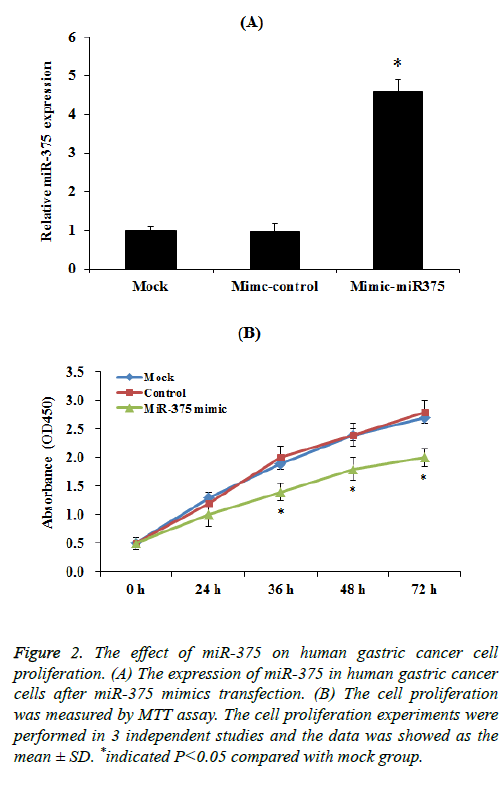
To select up-regulation of miR-375 expression, miR-375 mimic transfection assay was performed. Our results indicated that the expression of miR-375 was significantly increased compared to control group after mimic transfection (Figure 2A). Then we investigated the effect of miR-375 on gastric cancer cells proliferation. SGC-7901 cells transfected with a miR-375 mimic or miR-control were cultured for 72 h, the cell proliferation was measured by using the MTT assay every 24 h. MTT results demonstrated that the induction of miR-375 significantly inhibit the SGC-7901 cell proliferation especially 36 h after transfection (Figure 2B, p<0.05), while mimic control had no effect on SGC-7901 cell proliferation.
Figure 2: The effect of miR-375 on human gastric cancer cell proliferation. (A) The expression of miR-375 in human gastric cancer cells after miR-375 mimics transfection. (B) The cell proliferation was measured by MTT assay. The cell proliferation experiments were performed in 3 independent studies and the data was showed as the mean ± SD. *indicated P<0.05 compared with mock group.
Up-regulation of miR-375 induces gastric cancer cells apoptosis
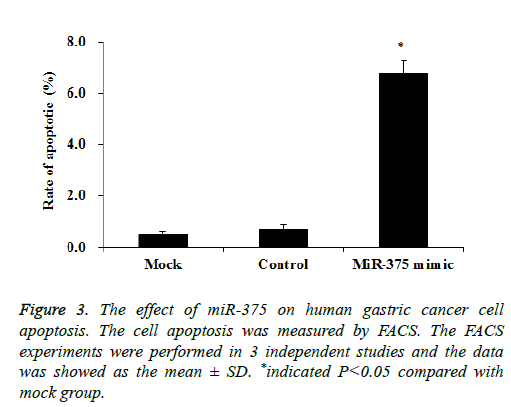
Then we tried to investigate whether the SGC-7901 cell proliferation inhibition was attributed to the cell apoptosis. SGC-7901 cells transfected with a miR-375 mimic or miRcontrol were cultured for 48 h, the cell apoptosis was measured by using the FACS assay. Our results showed transfection of miR-375 mimic significantly increased the SGC-7901 cell apoptosis to about 6.8% while it is only 0.6% in mock cells (p<0.05), while the control mimic did not affect the SGC-7901 cell apoptosis (Figure 3). Therefore miR-375 inhibit the SGC-7901 cell proliferation may be caused by the cell apoptosis induction.
MiR-375 targets AEG-1 in gastric cancer cells
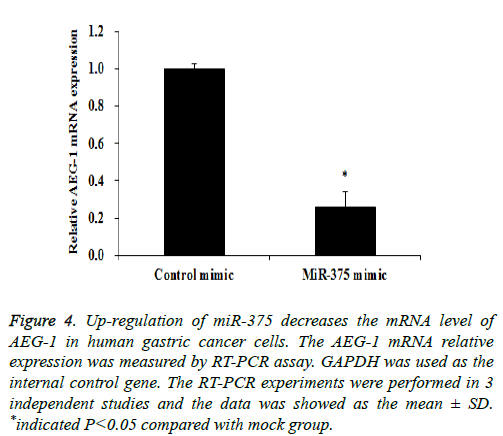
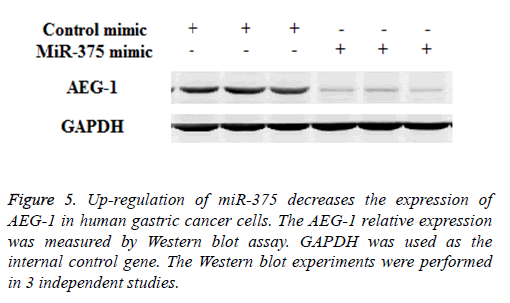
Previous study indicated that AEG-1 is a directed target of miR-375 [13], as a relatively novel gene, AEG-1 has emerged as an important oncogene in multiple aspects of cancer development and progression [14], in our study we try to find whether miR-375 regulate gastric cancer cell proliferation and apoptosis through targeting AEG-1 gene. To confirm our hypothesis Western Blot and RT-PCR assay were performed. Our RT-PCR results demonstrated that when up-regulation of miR-375, the AEG-1 mRNA level was significantly decreased (Figure 4) and the similar AEG-1 expression change also observed in protein level with Western Blot assay (Figure 5). Therefor miR-375 regulates the gastric cancer cell proliferation and apoptosis through the regulation of AEG-1 gene.
Figure 4: Up-regulation of miR-375 decreases the mRNA level of AEG-1 in human gastric cancer cells. The AEG-1 mRNA relative expression was measured by RT-PCR assay. GAPDH was used as the internal control gene. The RT-PCR experiments were performed in 3 independent studies and the data was showed as the mean ± SD. *indicated P<0.05 compared with mock group.
Discussion
Study indicated the abnormal expression miRs were closely associated with the progression of cancer and malignancy [15]. OncomiRs always high expressed in cancers and regulated the biological function of cancers, researchers reported that miR-10b was overexpressed in breast cancer and miR-21 was up-regulated in glioblastoma [16,17]; on the other hand tumor suppressor miRs were often down-regulated in cancers and they inhibit the carcinogenesis and progression of cancer, Tu et al. reported that miR-218 has been down-regulated in cancer and miR-218 regulated the cancer cell invasion, migration and proliferation [18].
Previous study indicated that miR-375 was first identified as an islet specific miRNA; it could regulate the insulin level [19], studies also demonstrated that the expression was downregulated in many type of cancers [9-11]. Study indicated that AEG-1 was an important oncogene in the development of cancer [14]; and AEG-1 was over-expressed in melanoma, glioma, neuroblastoma, and carcinomas of breast, prostate, esophagus, stomach and liver [20-27]. Previous study also found that AEG-1 was a directed target of miR-375 [13]; andd the relationship of the regulation of AEG-1 and miR-375 in gastric cancer has not yet been fully clarified. In the present study we found that the expression of miR-375 in gastric cancer cells was decreased compared to the normal gastric cells, induction of the miR-375 expression by mimic transfection could inhibit the gastric cancer cell proliferation and induce the gastric cancer cell apoptosis. Further study demonstrated miR-375 regulated the gastric cancer cell biological function through the regulation of AEG-1 gene.
Taken it together, our study suggested that miR-375 regulated the gastric cancer cell growth in vitro trough targeted AEG-1 gene in human gastric cancer cell, which could benefit for the gastric cancer clinical treatment. To better understand the relationship between the miR-375 and AEG-1 in gastric cancer cells, further studies are needed.
Acknowledgement
None
References
- Danaei G, Vander Hoorn S, Lopez AD, Murray CJ, Ezzati M; Comparative Risk Assessment collaborating group (Cancers). Causes of cancer in the world: comparative risk assessment of nine behavioural and environmental risk factors. Lancet 2005; 366: 1784-1793.
- Yasui W, Oue N, Kuniyasu H, Ito R, Tahara E. Molecular diagnosis of gastric cancer: present and future. Gastric Cancer 2001; 4: 113-121.
- Vauhkonen M, Vauhkonen H, Sipponen P. Pathology and molecular biology of gastric cancer. Best Pract Res Clin Gastroenterol 2006; 20: 651-674.
- Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol 2011; 223: 102-115.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281-297.
- Chitwood DH, Timmermans MC. Small RNAs are on the move. Nature 2010; 467: 415-419.
- Kosik KS. MicroRNAs and cellular phenotypy. Cell 2010; 143: 21-26.
- Tong F, Cao P, Yin Y, Xia S, Lai R. MicroRNAs in gastric cancer: from benchtop to bedside. Dig Dis Sci 2014; 59: 24-30.
- Avissar M, Christensen BC, Kelsey KT, Marsit CJ. MicroRNA expression ratio is predictive of head and neck squamous cell carcinoma. Clin Cancer Res 2009; 15: 2850-2855.
- Ewy AM, Giang HN, Elise DB, Yiqiang Z, Anuradha B, Aaron JS, Rosemary B, Mark R, Kensuke K, Duncan H, Nasser KA, Alan GC, Chang GL, Xin WW, Nozomu Y, Nobutoshi H, Andrew JD, Masao M, Carlo MC, Curtis CH. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res 2009; 15: 6192-6200.
- Liu AM, Poon RT, Luk JM. MicroRNA-375 targets Hippo-signaling effector YAP in liver cancer and inhibits tumor properties. Biochem Biophys Res Commun 2010; 394: 623-627.
- Smoot DT, Sewchand J, Young K, Desbordes BC, Allen CR, Naab T. A method for establishing primary cultures of human gastric epithelial cells. Methods Cell Sci 2000; 22: 133-136.
- He XX, Chang Y, Meng FY, Wang MY, Xie QH, Tang F, Li PY, Song YH, Lin JS. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene 2012; 31: 3357-3369.
- Hu G, Wei Y, Kang Y. The multifaceted role of MTDH/AEG-1 in cancer progression. Clin Cancer Res 2009; 15: 5615-5620.
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 2005; 353: 1793-1801.
- Parrella P, Barbano R, Pasculli B, Fontana A, Copetti M, Valori VM, Poeta ML, Perrone G, Righi D, Castelvetere M, Coco M, Balsamo T, Morritti M, Pellegrini F, Onetti-Muda A, Maiello E, Murgo R, Fazio VM. Evaluation of microRNA-10b prognostic significance in a prospective cohort of breast cancer patients. Mol Cancer 2014; 13: 142.
- Hermansen SK, Dahlrot RH, Nielsen BS, Hansen S, Kristensen BW. MiR-21 expression in the tumor cell compartment holds unfavorable prognostic value in gliomas. J Neurooncol 2013; 111: 71-81.
- Tu Y, Gao X, Li G, Fu H, Cui D, Liu H, Jin W, Zhang Y. MicroRNA-218 inhibits glioma invasion, migration, proliferation and cancer stem-like cell self-renewal by targeting the polycomb group gene Bmi. Cancer Res 2013; 73: 6046-6055.
- Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004; 432: 226-230.
- Kang DC, Su ZZ, Sarkar D, Emdad L, Volsky DJ. Cloning and characterization of HIV-1-inducible astrocyte elevated gene-1, AEG-1. Gene 2005; 353: 8-15.
- Lee SG, Jeon HY, Su ZZ, Richards JE, Vozhilla N. Astrocyte elevated gene-1 contributes to the pathogenesis of neuroblastoma. Oncogene 2009; 28: 2476-2484.
- Lee SG, Su ZZ, Emdad L, Sarkar D, Franke TF. Astrocyte elevated gene-1 activates cell survival pathways through PI3K-Akt signaling. Oncogene 2008; 27: 1114-1121.
- Li J, Zhang N, Song LB, Liao WT, Jiang LL, Gong LY, Wu J, Yuan J, Zhang HZ, Zeng MS, Li M. Astrocyte elevated gene-1 is a novel prognostic marker for breast cancer progression and overall patient survival. Clin Cancer Res 2008; 14: 3319-3326.
- Kikuno N, Shiina H, Urakami S, Kawamoto K, Hirata H, Tanaka Y, Place RF, Pookot D, Majid S, Igawa M, Dahiya R. Knockdown of astrocyte-elevated gene-1 inhibits prostate cancer progression through upregulation of FOXO3a activity. Oncogene 2007; 26: 7647-7655.
- Yu C, Chen K, Zheng H, Guo X, Jia W, Li M, Zeng M, Li J, Song L. Overexpression of astrocyte elevated gene-1 (AEG-1) is associated with esophageal squamous cell carcinoma (ESCC) progression and pathogenesis. Carcinogenesis 2009; 30: 894-901.
- Jian-bo X, Hui W, Yu-long H, Chang-hua Z, Long-juan Z. Astrocyte-elevated gene-1 overexpression is associated with poor prognosis in gastric cancer. Med Oncol 2011; 28: 455-462.
- Yoo BK, Emdad L, Su ZZ, Villanueva A, Chiang DY, Mukhopadhyay ND, Mills AS, Waxman S, Fisher RA, Llovet JM, Fisher PB, Sarkar D. Astrocyte elevated gene-1 regulates hepatocellular carcinoma development and progression. J Clin Invest 2009; 119: 465-477.