Research Article - Biomedical Research (2016) Volume 27, Issue 4
Transcriptome profiling identifies differentially expressed genes in systemic lupus erythematosus specific induced pluripotent stem cells from urine
Donge Tang1,2, Yuyu Chen2, Jianrong Huang2, Wujian Peng2, Kuibi Tan2, Honglei Wang2, Yong Dai*1Key Laboratory of Functional Protein Research of Guangdong Higher Education Institutes, Institute of Life and Health Engineering, College of Life Science and Technology, Jinan University, Guangzhou 510632, PR China
2Clinical Medical Research Center, the Second Clinical Medical College of Jinan University (Shenzhen People’s Hospital), Shenzhen, Guangdong, 518020, PR China
- *Corresponding Author:
- Yong Dai
Clinical Medical Research Center
The Second Clinical Medical College of Jinan University(Shenzhen People’s Hospital)
Shenzhen, PR China
Accepted date: April 17, 2016
Abstract
In clinical practice, it is difficult to monitor the repeating relapse in patients who have been suffering from systemic lupus erythematosus (SLE). The underlying etiology remains largely unknown. In order to understand the pathogenesis of SLE, the renal tubular cells-derived iPSCs were successfully obtained from urine of SLE patients. Here, with the purpose of identifying differentially expressed genes, highthroughput Illumina sequencing technology were analyzed the mRNA expression in SLE-iPSCs group and control-iPSCs group. Within the 4,254 genes, which were differentiated at least two-fold between the SLE-iPSCs and control-iPSCs, 2,856 genes were up-regulated and 1,398 down-regulated. These differentially expressed genes were involved in 9 cellular components, 9 molecular functions, 8 biological processes and 6 pathways with p-value ≤ 0.05. The clusters of “cellular process”, “intracellular” and “binding” represented the largest group in Process Ontology, Component Ontology, Function Ontology, respectively. Most differentially expressed genes involved in Function of binding, which were reported to be relevant with RNA transcription in SLE. Moreover, alternative splicing events and gene structure refinements of SLE-iPSCs group were greater than those of control-iPSCs group. Occurrence and development of SLE may be related to the excessive alternative spliced genes and events of alternative splicing. Using large cohorts of patient samples with long-term clinical follow-up data deserves for further investigation and research. Thus, it could assess the usefulness of the pathogenesis of SLE.
Keywords
Systemic lupus erythematosus-induced pluripotent stem cells (SLE- iPSCs), RNA sequence, Differentially expressed genes.
Introduction
Systemic lupus erythematosus (SLE) is the prototype of complex autoimmune disease characterized by the production of autoantibodies which results in widespread immunologic abnormalities and immune complex formation [1]. The patients can present variable manifestations and the nature courses are alternately remissions and relapses. Till now, although a lot of related researches have been undertaken [2], SLE patients still have no effective cures, whose treatments are often based upon long-term broad-spectrum immune suppressive regimes in the current therapeutic management. It has also become a major public health problem. Therefore, a rational approach for therapeutic design requires a detailed understanding of disease pathogenesis, and systematic characterization of the molecular and cellular basis of signaling abnormalities within the immune system and their relationship to regulation of gene expression remain critical for understanding [3].
The possibility of reprogramming somatic cells to induced pluripotent stem cells (iPSCs) offers an opportunity to generate pluripotent patient-specific cell lines, which can be conductive to studying pathogenesis of model human diseases [4]. Also, these iPSCs lines are powerful tools for drug discovery and the development of cellular transplantation therapies [4]. Generation of iPSCs from urine, fibroblasts and keratinocytes of disease patients has been reported [5-8]. To study SLE pathogenesis, renal tubular cells derived iPSCs were successfully established from urine of SLE patients [9]. In this study, high-throughput Illumina sequencing technology was utilized to further study the model cells of SLE-iPSCs.
Recently, the high-throughput Illumina Genome Analyzer provides a powerful approach to identify differentially expressed genes for the given cell, tissue and organism [10-12]. It can increase transcript sensitivity and identify novel transcripts, single nucleotide polymorphisms as well as splicing events [13]. Sequencing technology is developing rapidly, which results in the discovery of new disease genes. Actually, numerous chromosomal loci may harbor susceptibility genes [14,15]. By using the high-throughput Illumina sequencing platform, this study detects the mRNA expression in SLE-iPSCs group and control-iPSCs group, analyzes the two groups of differentially expressed genes and then identifies novel transcripts and splicing events. Further studies of gene regulation at transcriptional levels could help us to determine the pathogenesis of SLE comprehensively and accurately.
Materials and Methods
Materials
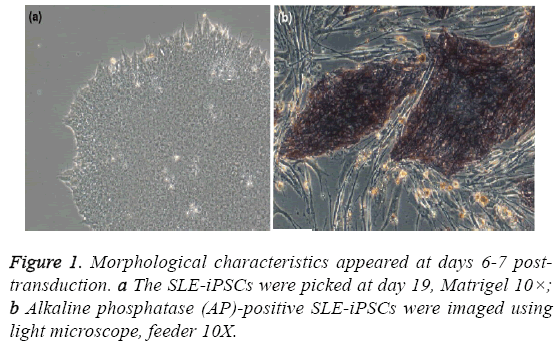
All the studies and other procedures were approved by the ethics committee of the Shenzhen People’s Hospital (Shenzhen, China) or the Guangzhou Institutes of Biomedicine and Health (Guangzhou, China). One patient, at the age of 39, was diagnosed as active SLE with SLEDAI>8. Moreover, one subject, age and sex matched, was recruited as healthy controls. To generate human iPSCs clone, renal tubular cells from urine of SLE patients were reprogrammed. Furthermore, SLE-iPSCs clone and control-iPSCs clone were identified [9]. Morphology was identified as well [9] (Figure 1). All of the samples were collected and immediately frozen in liquid nitrogen and stored at - 80°C.
RNA extraction, cDNA library construction and sequencing
The SLE-iPSCs and Control-iPSCs were utilized for RNA extraction, and total RNA of iPSCs was extracted as described previously [16]. The individual RNA samples were quantified and examined spectrophotometrically for protein contamination (A260/A280 ratio) and reagent contamination (A260/A230 ratio), which were prior to library construction.
The cDNA library was carried out at the Beijing Genomics Institute (BGI, Shenzhen, Guangdong, China) by using the Illumina manufacturer’s instructions. The main reagents and supplies were the Illumina Gene Expression Sample Prep Kit and the Solexa Sequencing Chip (flowcell), and the main instruments were the Illumina Cluster Station and the Illumina HiSeqTM 2000 System. In a nutshell, it is to extract the total RNA from samples. mRNA is enriched by utilizing the oligo (dT) magnetic beads and removing rRNA from the total RNA with kit. Through the fragmentation buffer, the mRNA is fragmented into short fragments (about 200~700 bp), Then, the first-strand cDNA is synthesized by random hexamer-primer using the mRNA fragments as templates. Buffer, dNTPs, RNase H and DNA polymerase I are added to synthesize the second strand cDNA. The double strand cDNA is purified with QiaQuick PCR extraction kit. Finally, sequencing adapters are ligated to the fragments. The fragments are purified by agarose gel electrophoresis and enriched by PCR amplification. The library products are ready for sequencing analysis via Illumina HiSeqTM 2000.
Bioinformatics analysis of sequencing data
Primary sequencing data produced by Illumina HiSeqTM 2000, called raw reads, is subjected to quality control (QC). To determine whether a resequencing step is necessary, the QC of alignment will be performed. Raw reads were filtered to remove reads with adapters, unknown bases of more than 10% and low quality reads (which are defined as reads having more than 50% bases with quality value ≤ 10). After filtering, the remaining reads, which are called “clean reads”, will be aligned to the reference sequences with SOAP2 [17]. Then, QC of alignment is performed again. The alignment data is utilized to calculate the distribution of reads on reference genes and perform coverage analysis. If alignment results pass QC, we will proceed with downstream analysis including gene expression, gene structure refinement, alternative splicing, novel transcript prediction and SNP detection.
Gene expression analysis
Results of gene expression contain gene expression levels and differential expression analysis. The levels of gene expression were measured as numbers of reads per kilobase of exon model in a gene per million mapped reads (RPKM), which can be directly utilized to compare the differences of gene expression among the samples.
According to Differentially Expressed Genes (DEGs), we utilize ‘FDR(false discovery rate) ≤ 0.001 [18] and the absolute value of log2-Ratio ≥ 1 ’as the thresholds to judge the significance of DEGs. Then, more stringent criteria with smaller FDR and bigger fold change value can be applied to identify DEGs. These Differentially Expressed Genes were annotated in Gene Ontology (GO) and KEGG pathway analysis.
GO enrichment analysis maps all DEGs to GO terms in the database (http://www.geneontology.org/), calculating gene numbers for each term. Beyond that, the calculated p-value goes through Bonferroni Correction [19], taking corrected pvalue ≤ 0.05 as a threshold. GO terms fulfilling this condition are defined as significantly enriched GO terms in DEGs. This analysis can recognize the main biological functions exercised by DEGs.
Result
High-throughput sequencing and reads mapping
In order to identify SLE-related genes, transcript profiling of iPSCs was performed using RNA-Seq. Clean reads are mapped to genome sequence by using SOAP2 aligner.
Through mapping of reads to entire genome sequence, a total of 26,749,056 and 26,855,414 high-quality reads were obtained for the SLE-iPSCs and the control-iPSCs, respectively (Table 1). After discarding the low quality tags, 2,407,415,040 and 2,416,987,260 tags remained in the SLE-iPSCs and controliPSCs, respectively. 23,488,075 (87.81%) and 23,928,072 (89.10%) reads were mapped to the whole genome, 16,513,998 (61.74%) and 15,718,886 (58.53%) reads were perfect match in the SLE-iPSCs and control-iPSCs, respectively. 795,112 reads were expressed exclusively in the SLE-iPSCs, which were possibly related to the SLE development. Further analysis revealed that 21,382,507 unique tags (79.94%) and 21,937,249 (81.69%) were exclusively matched in the SLE-iPSCs and control-iPSCs, respectively. These data indicate that approximately 80% of the transcripts are expressed in the SLEiPSCs and control-iPSCs (Table 1).
| Map to Genome | Total Reads | Total BasePairs | Total Mapped Reads | perfect match | <=5bp mismatch | unique match | multi- position match | Total Unmapped Reads |
|---|---|---|---|---|---|---|---|---|
| SLE- iPSC | 26749056 | 2407415040 | 23488075 | 16513998 | 6974077 | 21382507 | 2105568 | 3260981 |
| (%) | 100.00 | 100.00 | 87.81 | 61.74 | 26.07 | 79.94 | 7.87 | 12.19 |
| Control- iPSC | 26855414 | 2416987260 | 23928072 | 15718886 | 8209186 | 21937249 | 1990823 | 2927342 |
| (%) | 100.00 | 100.00 | 89.10 | 58.53 | 30.57 | 81.69 | 7.41 | 10.90 |
Table 1: The SLE-iPSC and control-iPSC reads were mapped to the human Genome.
Gene structure refinement
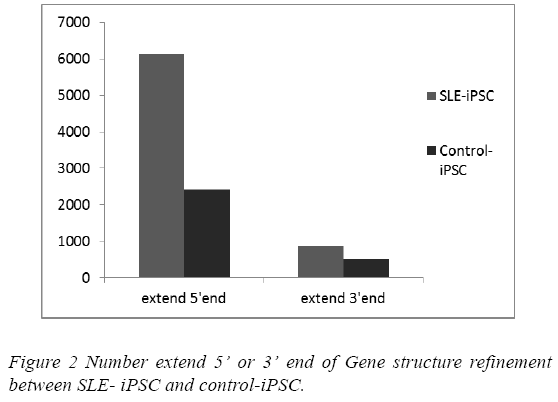
Mapping to genome allows for potential discovery of novel exons and novel 5’ and 3’ ends. This can increase sensitivity and specificity due to recognition of potential reads through transcripts from neighboring genes. Transcripts were assembled with reads by Cufflinkss [20]. Through comparing assembled transcripts and gene annotation from reference sequences, it’s possible to find assembled transcripts that can extend 5’ or 3’ end of gene annotation, and therefore refine gene structure. 6,985 and 2,935 gene structures were refined in the SLE-iPSCs and control-iPSCs, respectively. Gene structure refinements in the SLE-iPSCs were apparently higher than those in the control-iPSCs (Figure 2).
Novel transcript regions were also detected using Cufflinkss. As a novel transcript, an assembled transcript must meet three requirements. This means that the transcript must be at least 200 bp away from annotated gene, the transcript is of length over 180 bp and the sequencing depth is no less than 2. In our experiment, 792 and 973 novel transcribed regions were found in the SLE-iPSCs and control-iPSCs, respectively.
Alternative splicing analysis
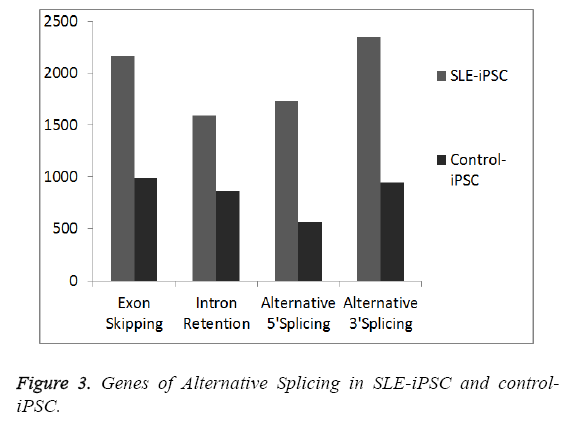
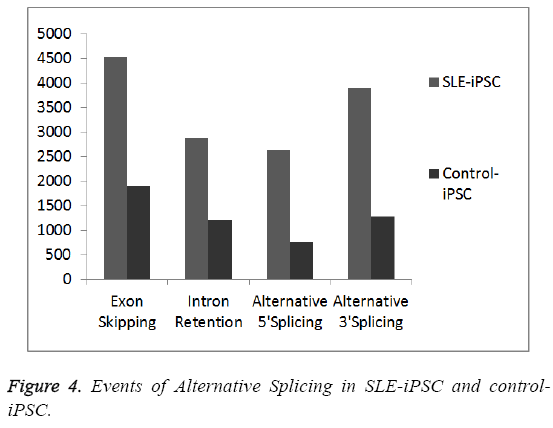
To generate a mature mRNA, the introns must be removed and exons joined together in the process of pre-mRNA splicing [21]. Many pre-mRNAs in humans and other metazoans undergo the process of alternative splicing (AS) where multiple mRNA isoforms are produced from a single gene locus. In order to detect gene junctions, we perform an “intact” alignment using SOAPsplice to map complete reads to the reference genome. The remaining reads, initially unmapped reads (IUM reads), are mapped with the spliced alignment algorithm. We mainly detect four kinds of alternative spliced events, exon skipping, intron retention, alternative 5’ splice site and alternative 3’ splice site (Figures 3 and 4). Our results indicate that SLE-iPSCs have a higher rate of alternative splicing, compared with the control-iPSCs (Figure 4). And alternatively spliced genes of SLE-iPSCs are significantly higher than those of control-iPSCs (Figure 3). Disruption of normal splicing or splicing misregulation has been observed in a large number of diseases, which has been reviewed extensively [22-24]. The results clearly show that different organisms have different levels of alternative splicing, as well as alternatively spliced genes. Occurrence and development of SLE may be related to the excessive alternatively spliced genes and events of alternative splicing.
Global analysis of gene expression
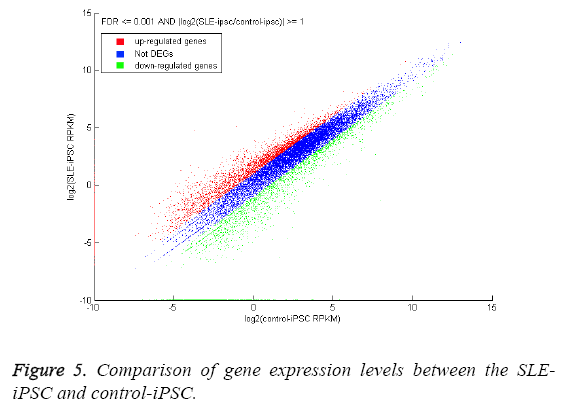
One of the primary goals of transcriptome sequencing is to compare gene expression levels in different genotypes. We estimated the expression levels of genes between the SLEiPSCs and the control-iPSCs. The expressed levels are measured in “reads per kilobase of exon model per million mapped reads” (RPKM), and the expression level of one gene is the sum of the RPKM values of its isoforms [25]. A total of 18,142 annotated genes were detected with RPKM>0. Using the P-value ≤ 0.05 and FDR ≤ 0.001 as threshold value, 4,254 genes were detected at least two-fold difference between the SLE-iPSCs and control-iPSCs (Figure 5). 2,856 genes and 1,398 genes represent a higher and lower abundance of more than two fold than those of control-iPSCs, respectively. The blue dots representing differentially expressed genes are less than two-fold between the two libraries, which are arbitrarily designated as ‘‘no difference in expression’’. We observe that parts of the dots are on the axis. Red dots of Y-axis indicate that genes only express in SLE-iPSCs, and green dots of Xaxis only express in control-iPSCs. The differentially expressed genes with ten-fold or greater are shown in Tables 2 and 3. We find that most differential expressed genes are involved in binding, ion binding, GTPase regulator activity, nucleoside- triphosphatase regulator activity of Function Ontology (Table 3) and so on.
| Gene ID | Description | Log2ratio | GO Function |
|---|---|---|---|
| 7503 | X (inactive)-specific transcript (non-protein coding) | 13.92469 | - |
| 647135 | SLIT-ROBO Rho GTPase activating protein 2 pseudogene 2 | 13.37934 | binding; enzyme activator activity |
| 400680 | hypothetical LOC400680 | 12.16684 | - |
| 728739 | programmed cell death 2 pseudogene | 11.70369 | metal ion binding |
| 391322 | D-dopachrometautomerase-like | 11.67755 | binding;carboxy-lyase activity; intramolecularoxidoreductase activity |
| 8577 | transmembrane protein with EGF-like and two follistatin-like domains 1 | 11.37174 | - |
| 128486 | fat storage-inducing transmembrane protein 2 | 11.08058 | - |
| 343171 | olfactory receptor, family 2, subfamily W, member 3 | 11.05965 | G-protein coupled receptor activity |
| 645455 | centrosomal protein 170kDa pseudogene 1 | 11.04503 | - |
| 3043 | hemoglobin, beta | 10.84645 | iron ion binding;protein binding |
| 27023 | forkhead box B1 | 10.82734 | sequence-specific DNA binding RNA polymerase II transcription factor activity |
| 168620 | basic helix-loop-helix family, member a15 | 10.74333 | nucleic acid binding;identical protein binding |
| 285598 | ADP-ribosylation factor-like 10 | 10.73247 | nucleotide binding |
| 85439 | stonin 2 | 10.58868 | binding |
| 5456 | POU class 3 homeobox 4 | 10.427 | nucleic acid binding transcription factor activity;sequence-specific DNA binding;structure-specific DNA binding |
| 3188 | heterogeneous nuclear ribonucleoprotein H2 (H') | 10.34543 | poly-pyrimidine tract binding |
| 642968 | family with sequence similarity 163, member B | 10.31501 | - |
| 80108 | zinc finger protein 2 homolog | 10.31064 | nucleic acid binding transcription factor activity;nucleic acid binding;transition metal ion binding |
| 339535 | hypothetical LOC339535 | 10.13272 | - |
| 56134 | protocadherin alpha subfamily C, 2 | 10.12553 | signal transducer activity;metal ion binding |
| 2949 | glutathione S-transferase mu 5 | 10.09299 | transferase activity |
| 10887 | prokineticin receptor 1 | 10.06696 | neuropeptide receptor activity |
| 27091 | calcium channel, voltage-dependent, gamma subunit 5 | 10.04713 | calcium channel activity |
Table 2: Up-regulated of the differential expressed genes with ten-fold or greater.
| Gene ID | Description | Log2ratio | GO Function |
|---|---|---|---|
| 9086 | eukaryotic translation initiation factor 1A, Y-linked | -15.3561 | translation factor activity, nucleic acid binding |
| 8653 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | -13.7175 | nucleic acid binding;RNA helicase activity;ATPase activity, coupled |
| 55410 | non-protein coding RNA 185 | -13.6707 | - |
| 84663 | chromosome Y open reading frame 15B | -13.5977 | - |
| 22829 | neuroligin 4, Y-linked | -12.719 | binding;identical protein binding; |
| 5616 | protein kinase, Y-linked | -12.7115 | cyclic nucleotide-dependent protein kinase activity; |
| 256536 | transcription elongation regulator 1-like | -12.603 | binding |
| 83869 | testis-specific transcript, Y-linked 14 | -12.454 | - |
| 8284 | lysine (K)-specific demethylase 5D | -12.052 | nucleic acid binding;oxidoreductase activity;transition metal ion binding |
| 246126 | chromosome Y open reading frame 15A | -12.0162 | - |
| 6736 | sex determining region Y | -12.0071 | nucleic acid binding transcription factor activity;nucleic acid binding;transcription regulator activity |
| 9506 | P antigen family, member 4 | -11.254 | - |
| 64595 | testis-specific transcript, Y-linked 15 | -11.253 | sequence-specific DNA binding |
| 728640 | family with sequence similarity 133, member B pseudogene | -10.9706 | - |
| 9087 | thymosin beta 4, Y-linked | -10.9171 | cytoskeletal protein binding |
| 729609 | hypothetical LOC729609 | -10.9048 | binding |
| 8287 | ubiquitin specific peptidase 9, Y-linked | -10.6199 | endopeptidase activity;thiolester hydrolase activity;small conjugating protein-specific protease activity;SMAD binding |
| 130367 | sphingosine-1-phosphate phosphatase 2 | -10.456 | phosphatase activity |
| 1419 | crystallin, gamma B | -10.4233 | structural molecule activity |
| 93408 | myosin, light chain 10, regulatory | -10.266 | metal ion binding |
| 7404 | ubiquitously transcribed tetratricopeptide repeat gene, Y-linked | -10.2028 | oxidoreductase activity;cation binding |
| 56891 | lectin, galactoside-binding, soluble, 14 | -10.1905 | carboxylesterase activity;hydrolase activity;carbohydrate binding |
| 26212 | olfactory receptor, family 2, subfamily B, member 6 | -10.1134 | G-protein coupled receptor activity |
| 100128124 | hypothetical LOC100128124 | -10.1112 | - |
| 376693 | ribosomal protein S10 pseudogene 7 | -10.0133 | structural molecule activity;binding |
Table 3: Down-regulated of the differential expressed genes with tenfold or greater.
Gene ontology functional enrichment analysis of differentially expressed genes (DEGs)
Gene Ontology (GO) is an international standardized gene functional classification system, which offers a dynamic update and a strictly defined concept to comprehensively describe the properties of genes and their products in any organism.
Using the P-value ≤ 0.05 as the threshold value, 4,254 differentially expressed genes were categorized into 30 functional groups (Tables 4-6), which included 9 molecular functions (Table 4), 9 cellular components (Table 5) and 8 biological processes (Table 6).
| Accession | Gene Ontology term | Numbers of DEG involved in term | Numbers of DEG involved in ontology | Cluster frequency | Corrected P-value |
| GO:0005488 | binding | 2825 | 3274 | 86.30% | 2.28E-08 |
| GO:0043167 | ion binding | 997 | 3274 | 30.50% | 5.76E-13 |
| GO:0043169 | cation binding | 986 | 3274 | 30.10% | 5.43E-13 |
| GO:0046872 | metal ion binding | 808 | 3274 | 24.70% | 2.27E-12 |
| GO:0046914 | transition metal ion binding | 552 | 3274 | 16.90% | 5.15E-06 |
| GO:0030695 | GTPase regulator activity | 117 | 3274 | 3.60% | 0.00012 |
| GO:0060589 | nucleoside-triphosphatase regulator activity | 118 | 3274 | 3.60% | 9.65E-05 |
| GO:0005085 | guanyl-nucleotide exchange factor activity | 36 | 3274 | 1.10% | 0.02771 |
| GO:0005088 | Rasguanyl-nucleotide exchange factor activity | 36 | 3274 | 1.10% | 0.0159 |
Table 4: The classification of differentially expressed genes in different category based on function ontology.
| Accession | Gene Ontology term | Numbers of DEG involved in term | Numbers of DEG involved in ontology | Cluster frequency | Corrected P-value |
| GO:0005622 | intracellular | 2510 | 3350 | 74.90% | 0.00016 |
| GO:0044424 | intracellular part | 2497 | 3350 | 74.50% | 0.00013 |
| GO:0043226 | organelle | 2140 | 3350 | 63.90% | 0.00358 |
| GO:0043229 | intracellular organelle | 2115 | 3350 | 63.10% | 0.01626 |
| GO:0044422 | organelle part | 1056 | 3350 | 31.50% | 0.01696 |
| GO:0005634 | nucleus | 461 | 3350 | 13.80% | 0.03414 |
| GO:0044428 | nuclear part | 449 | 3350 | 13.40% | 0.03634 |
| GO:0005840 | ribosome | 61 | 3350 | 1.80% | 0.03914 |
| GO:0005840 | ribosomal subunit | 58 | 3350 | 1.70% | 0.00816 |
Table 5: The classification of differential expressed genes in different category based on component ontology.
| Accession | Gene Ontology term | Numbers of DEG involved in term | Numbers of DEG involved in ontology | Cluster frequency | Corrected P-value |
|---|---|---|---|---|---|
| GO:0009987 | cellular process | 2264 | 3067 | 73.80% | 0.00099 |
| GO:0043170 | macromolecule metabolic process | 1154 | 3067 | 37.60% | 0.01762 |
| GO:0044260 | cellular macromolecule metabolic process | 1005 | 3067 | 32.80% | 3.27E-05 |
| GO:0007275 | multicellular organismal development | 661 | 3067 | 21.60% | 0.02366 |
| GO:0044267 | cellular protein metabolic process | 568 | 3067 | 18.50% | 8.92E-05 |
| GO:0007155 | cell adhesion | 143 | 3067 | 4.70% | 0.00025 |
| GO:0022610 | biological adhesion | 143 | 3067 | 4.70% | 0.00025 |
| GO:0016337 | cell-cell adhesion | 94 | 3067 | 3.10% | 3.27E-05 |
Table 6: The classification of differential expressed genes in different category based on process ontology.
According to biological process, 3,067 differentially expressed genes were involved in biological process. The genes involved in cellular process (2264) [GO: 0009987] were enriched more significantly than other seven biological processes. The next representative terms were metabolic process (macromolecule metabolic process, cellular macromolecule metabolic process, cellular protein metabolic process) (Table 6). According to cellular component, 3,350 differentially expressed genes were involved in cellular component. The most representative GO term was intracellular (GO: 0005622), the next representative GO term was organelle (GO: 0043226), which was carried out at the intracellular level and results in the biosynthesis of constituent macromolecules, assembly, arrangement of constituent parts, or disassembly of an intracellular component (Table 5). The significantly enriched transcripts on the Function Ontology were 2,825 DEGs, which associated with the GO term of binding [GO: 0005488], the differentially expressed genes were also involved in ion binding, cation binding, metal ion binding, transition metal ion binding. The remaining of differentially expressed genes was categorized into regulator activity (GTPase regulator activity, nucleosidetriphosphatase regulator activity) and exchange factor activity (guanyl-nucleotide exchange factor activity, Ras guanylnucleotide exchange factor activity). Alessandra B and Pernis reported that deregulation of Rho GTPase-mediated pathways might play a role in the pathogenesis of SLE [26].
Pathway enrichment analysis for DEGs
Genes usually interact with each other in certain biological functions. The Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway database contains a record of all known networks of molecular interactions in cells and the variants of these pathways that are specific to particular organisms [27]. Pathway enrichment analysis helps us to understand what biological functions the genes have and how these genes interact with each other. In this study, the KEGG database can be used to analyze the potential involvement of differentially expressed genes.
Pathways with Q-value ≤ 0.05 were significantly enriched in differentially expressed genes. The differentially expressed genes were categorized into six pathways. The most representative pathway is purine metabolism, and then 4.42% of differentially expressed genes were involved in pathways in cancer (Table 7). Purinergic signaling not only regulated numerous organ systems, but also involved in embryonic development, injury and pain [28]. Moreover, there is evidence that purinergic receptors play a role in the regulation of behaviours and immunity [29]. Our result shows most significant difference of expressed genes in SLE-iPSCs involve in purine metabolism.
| Pathway ID | Pathway | Cluster frequency | P-value | Q-value |
| ko00230 | Purine metabolism | 9.27% | 0.000848 | 0.033532 |
| ko05200 | Pathways in cancer | 4.42% | 0.00021 | 0.016019 |
| ko05222 | Small cell lung cancer | 1.48% | 0.000173 | 0.016019 |
| ko03010 | Ribosome | 1.42% | 7.07E-05 | 0.016019 |
| ko04350 | TGF-beta signaling pathway | 1.40% | 0.000879 | 0.033532 |
| ko05220 | Chronic myeloid leukemia | 1.34% | 0.000286 | 0.016353 |
Table 7: Pathway enrichment analysis for DEGs.
Discussion
During the past few years, new NGS technologies have been developed with applications in complete genome sequencing, metagenomic sequencing, Chip-Seq, small RNA sequencing, transcriptome profiling, and others. Yukinori Okada et al [30] uses genome-wide association studies (GWAS) to discovery of susceptibility genes of SLE. And Graham, Deborah S Cunninghame [31] indicates that GWAS has been shown to be a powerful way of identifying novel susceptibility genes in SLE.
Our study provides the first comprehensive insight into the transcriptome of SLE- iPSCs using Illumina HiSeqTM 2000, as a powerful next generation RNA sequencing platform. We calculated the number of differentially expressed genes between SLE- iPSCs and control-iPSCs based on RPKM. 4,254 genes were considered to be significant difference, classified using Gene Ontology (GO) and KEGG Pathway. The clusters of “cellular process”, “intracellular” and “binding” represented the largest group in Process Ontology, Component Ontology and Function Ontology, respectively. Most differentially expressed genes involved in Function of binding (Tables 2 and 3), such as up-regulated hemoglobin, forkhead box B1, basic helix-loop-helix family and down- regulated eukaryotic translation initiation factor 1A, transcription elongation regulator 1-like and so on. Most of them were reported to be relevant with RNA transcription in SLE [32]. Bhatnagar et al. [33] showed that human Hemoglobin (Hb) was demonstrated in the sera of systemic lupus erythematosus (SLE). In addition, heterogeneous nuclear ribonucleoprotein H2 was significant difference in the study, which was consistent with our previous results [34]. Siapka et al. also demonstrated that hnRNP had a predominant nuclear localization and exerted multiple functions, including regulation of alternative splicing, transport of mRNA and regulation of translation [35].
Alternative splicing has recently emerged as a major mechanism for expanding and regulating the repertoire of gene functions [36]. Alternatively spliced proteins are involved in many biological processes like apoptosis [37]. The effects of these polymorphisms on splicing efficiency are believed to contribute significantly to disease severity and susceptibility [38]. Most alternative splicing events affect the coding sequence, subsequently amino acid sequence. Recently, a number of observations indicate physiological or pathological significance of alternative splicing and its role in the genetic background of diseases [39], such as immune diseases [40]. About one third of AS events result in splice variants containing premature termination codon, or leading to nonsense-mediated decay of the RNA product [41]. Many disease- associated mutations also affect pre-mRNA alternative splicing, usually causing inappropriate exon skipping [42]. Our result indicates that four types of exon skipping, intron retention, alternative 5’ splice site and alternative 3’ splice site in SLE-iPSCs are higher than those in control-iPSCs. Exon skipping in SLE-iPSCs is the biggest difference among the four types of alternatives. Studies have shown that up to 50% of point mutations responsible for genetic diseases in humans cause aberrant splicing [43,44]. The most common phenotype of point mutations that affect splicing, however, is exon skipping [43,45]. A family-based analysis of Caucasian and Chinese populations shows a significant association between the major alleles of a three alternative splicing CR2 and lupus susceptibility [46]. KB Douglas et al. also confirm the association of these three CR2 variants, and identify two additional CR2 variants significantly associated with SLE susceptibility. Therefore, aberrant exon skipping is closely related to the pathogenesis of SLE, and as the most common phenotype of alternatives because it is not usually amenable to correction.
Conclusion
The high-throughput Illumina sequencing technology is a powerful approach to identification of differentially expressed genes. We used the Illumina sequencing technology to detect the mRNAs expression in SLE-iPSCs group and control-iPSCs group; 4,254 genes were detected at least two-fold difference between the SLE-iPSCs and control-iPSCs, 2,856 genes were up-regulated and 1,398 down-regulated. Some differentially expressed genes only express in SLE-iPSCs or control-iPSCs. The 4,254 differentially expressed genes were annotated in Gene Ontology (GO) and KEGG pathway analysis. We found that the DEGs involved in 9 cellular components, 9 molecular functions, 8 biological processes and 6 pathways with p-value ≤ 0.05. The clusters of “cellular process”, “intracellular” and “binding” represented the largest group in Process Ontology, Component Ontology, Function Ontology, respectively. Most differentially expressed genes involved in Function of binding, which were reported to be relevant with RNA transcription in SLE. Moreover, we also proceeded with other downstream analysis including gene structure refinement, alternative splicing and novel transcript prediction. Alternative splicing events and gene structure refinement of SLE-iPSCs group were greater than those of control-iPSCs group. The results clearly showed that different organisms had different levels of alternative splicing, as well as alternatively spliced genes. Occurrence and development of SLE may be related to the excessive alternatively spliced genes and events of alternative splicing. In the future, further investigation is necessary for using large cohorts of patient samples with long-term clinical follow-up data, to assess the usefulness of the pathogenesis of SLE.
Acknowledgements
We thank the patients and healthy volunteers who participated in this study.
Conflict of Interest
No financial relationship exists between any of the authors and any commercial interest with a vested interest in the outcome of the study.
References
- Perl A. Pathogenic mechanisms in systemic lupus erythematosus. Autoimmunity 2009; 43:1-6.
- de Macedo PA, Borba EF, VianaVdST, Leon EP, TestagrossaLdA, Barros RT, Nascimento AP, Bonfá E. Antibodies to ribosomal P proteins in lupus nephritis: A surrogate marker for a better renal survival. Autoimmunity reviews 2011; 10:126-130.
- Kyttaris VC, Krishnan S, Tsokos GC. Systems biology in systemic lupus erythematosus: integrating genes, biology and immune function. Autoimmunity 2006; 39:705-709.
- Malik N, Rao MS. A review of the methods for human iPSC derivation. In: Pluripotent Stem Cells. Springer; 2013; 23-33.
- Guan X, Mack DL, Moreno CM, Strande JL, Mathieu J, Shi Y, Markert CD, Wang Z, Liu G, Lawlor MW. Dystrophin-deficient cardiomyocytes derived from human urine: New biologic reagents for drug discovery. Stem cell research 2014; 12:467-480.
- Thatava T, Armstrong AS, De Lamo JG, Edukulla R, Khan YK, Sakuma T, Ohmine S, Sundsbak JL, Harris PC, Kudva YC. Successful disease-specific induced pluripotent stem cell generation from patients with kidney transplantation. Stem Cell Res Ther 2011; 2:48.
- Koh S, Piedrahita JA. Generation of Induced Pluripotent Stem Cells (iPSCs) from Adult Canine Fibroblasts. Methods in molecular biology 2015; 1330:69-78.
- Hung SSC, Pebay A, Wong RCB: Generation of Integration-free Human Induced Pluripotent Stem Cells Using Hair-derived Keratinocytes. Jove-J Vis Exp2015; 102.
- Chen Y, Luo R, Xu Y, Cai X, Li W, Tan K, Huang J, Dai Y. Generation of systemic lupus erythematosus-specific induced pluripotent stem cells from urine. Rheumatology international 2013; 33:2127-2134.
- Feero WG, Guttmacher AE, Manolio TA.Genomewide association studies and assessment of the risk of disease. New England Journal of Medicine 2010; 363:166-176.
- Han X, Wu X, Chung W-Y, Li T, Nekrutenko A, Altman NS, Chen G, Ma H.Transcriptome of embryonic and neonatal mouse cortex by high-throughput RNA sequencing. Proceedings of the National Academy of Sciences 2009; 106:12741-12746.
- Ozer A, Tome JM, Friedman RC, Gheba D, Schroth GP, Lis JT. Quantitative assessment of RNA-protein interactions with high-throughput sequencing-RNA affinity profiling. Nature protocols 2015; 10:1212-1233.
- Marguerat S, Bähler J. RNA-seq: from technology to biology. Cellular and molecular life sciences 2010; 67:569-579.
- Coonrod EM, Durtschi JD, Margraf RL, Voelkerding KV. Developing genome and exome sequencing for candidate gene identification in inherited disorders: an integrated technical and bioinformatics approach. Archives of pathology & laboratory medicine 2013; 137:415-433.
- Tian Y, Wang D. Analysis of susceptible genes and chromosome loci for lung cancers by automated gene prediction tools and genome scanning meta-analysis. International journal of clinical and experimental pathology 2015; 8:11929-11933.
- Li JB, Levanon EY, Yoon J-K, Aach J, Xie B, LeProust E, Zhang K, Gao Y, Church GM. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 2009; 324:1210-1213.
- Li R, Yu C, Li Y, Lam T-W, Yiu S-M, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 2009; 25:1966-1967.
- Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Annals of statistics 2001;1165-1188.
- Salkind NJ.Encyclopedia of measurement and statistics, Sage Publications, 2007.
- Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 2011; 27:2325-2329.
- Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nature Reviews Molecular Cell Biology 2005; 6:386-398.
- Baralle D, Baralle M. Splicing in action: assessing disease causing sequence changes. Journal of medical genetics 2005; 42:737-748.
- Wang G-S, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nature Reviews Genetics 2007; 8:749-761.
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell 2009; 136:777-793.
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods 2008; 5:621-628.
- Pernis AB. Rho GTPase-mediated pathways in mature CD4T cells. Autoimmunity reviews 2009; 8:199-203.
- Xu P, Liu Z, Fan X, Gao J, Zhang X, Zhang X, Shen X. De novotranscriptome sequencing and comparative analysis of differentially expressed genes inGossypiumaridumunder salt stress. Gene 2013; 525:26-34.
- Koles L, Furst S, Illes P. Purine ionotropic (P2X) receptors. Current pharmaceutical design 2007; 13:2368-2384.
- Kim DS, Palmiter RD. Interaction of dopamine and adenosine receptor function in behavior: studies with dopamine-deficient mice. Frontiers in bioscience: a journal and virtual library 2008; 13:2311.
- Okada Y, Shimane K, Kochi Y, Tahira T, Suzuki A, Higasa K, Takahashi A, Horita T, Atsumi T, Ishii T. A genome-wide association study identified AFF1 as a susceptibility locus for systemic lupus eyrthematosus in Japanese. PLoS genetics 2012; 8:e1002455.
- Graham DSC. Genome-wide association studies in systemic lupus erythematosus: a perspective. Arthritis research & therapy 2009; 11:119.
- Braunstein I, Klein R, Okawa J, Werth V. The interferon-regulated gene signature is elevated in subacute cutaneous lupus erythematosus and discoid lupus erythematosus and correlates with the cutaneous lupus area and severity index score. British Journal of Dermatology 2012; 166:971-975.
- Bhatnagar H, Kala S, Sharma L, Jain S, Kim KS, Pal R. Serum and organ-associated anti-hemoglobinhumoralautoreactivity: Association with anti-Sm responses and inflammation. European journal of immunology 2011; 41: 537-548.
- Sui W, Tang D, Zou G, Chen J, Ou M, Zhang Y, Dai Y. Differential proteomic analysis of renal tissue in lupus nephritis using iTRAQ reagent technology. Rheumatology international 2012; 32:3537-3543.
- Siapka S, Patrinou-Georgoula M, Vlachoyiannopoulos PG, Guialis A. Multiple specificities of autoantibodies against hnRNP A/B proteins in systemic rheumatic diseases and hnRNP L as an associated novel autoantigen. Autoimmunity 2007; 40:223-233.
- Graveley BR. Alternative splicing: increasing diversity in the proteomic world. TRENDS in Genetics 2001; 17:100-107.
- Schwerk C, Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Molecular cell 2005; 19:1-13.
- Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nature reviews Genetics 2007; 8:749-761.
- Pagani F, Baralle FE. Genomic variants in exons and introns: identifying the splicing spoilers. Nature reviews Genetics 2004; 5:389-396.
- Amano MT, Ferriani VPL, Florido MPC, Reis ES, Delcolli MIMV, Azzolini AECS, Assis-Pandochi AI, Sjöholm AG, Farah CS, Jensenius JC. Genetic analysis of complement C1s deficiency associated with systemic lupus erythematosus highlights alternative splicing of normal C1s gene. Molecular Immunology 2008; 45:1693-1702.
- Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proceedings of the National Academy of Sciences 2003; 100:189-192.
- Cartegni L, Krainer AR. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nature structural biology 2003; 10:120-125.
- Luca C, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Reviews Genetics 2002; 3:285-298.
- Ars E, Serra E, García J, Kruyer H, Gaona A, Lázaro C, Estivill X. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Human Molecular Genetics 2000; 9:237-247.
- Nakai K, Sakamoto H. Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene 1994; 141:171-177.