Research Article - Journal of Genetics and Molecular Biology (2021) Volume 5, Issue 4
Thirteen Polymorphic STR Loci in the HLA Region: can they Predict HLA Alleles in South Tunisia?.
Nadia Mahfoudh*, Adia Charfi, Lilla Gaddour, Faiza Hakim, Hafedh Makni, Arwa KamounDepartment of Histocompatibility, Hedi Chaker Hospital, Faculty of Medicine, University of Sfax, Sfax, Tunisia
- *Corresponding Author:
- Nadia Mahfoudh
Department of Histocompatibility
Hedi Chaker Hospital, Faculty of Medicine
University of Sfax
Sfax, Tunisia
Tel: +21698445245
E-mail: Mahfoudh.nadia@yahoo.fr
Accepted on June 29, 2021
Abstract
In the HLA region, several microsatellites (Msats) also called Short Tandem Repeats (STR) were mapped. Msats are not themselves functional; however, their inherent polymorphism and linkage disequilibrium (LD) with HLA loci make them a robust disease-mapping tool in understanding susceptibility to autoimmune and infectious diseases. The aims of our study were to define a set of 13 STRs were evenly distributed in the HLA, to evaluate their LD with HLA alleles; and to test Msats ability to predict HLA typing. HWE was verified for all STRs except the TNFb and D6S1666 Msats. Regarding statistical parameters, we used LD and HSH analysis to ascertain the best MSAts for HLA prediction. A marker in strong LD with an HLA locus and with a low value of HSH is the most appropriate for predicting HLA alleles. For the HLA-A1-B52-DR15 haplotype, the combination of the alleles markers D6S265 (a10), D6S2810 (a7), STR-MICA (a6) and D6S2789 (a16) was necessary for haplotype prediction. In conclusion, for prediction accuracy we found that the positive predictive value (PPV), the probability of observing the particular HLA haplotype in the presence of a particular Msats allele, was the most relevant statistical parameter.
Keywords
HLA, Microsatellites, STR, Prediction.
Introduction
In the HLA region, several microsatellites (Msats) also called Short Tandem Repeats (STR) were mapped. It is estimated that there is a Msat in the Human Leucocytes Antigen (HLA) region every 30 kb [1]. In September 1999 [2], 155 polymorphic Msats were identified and published. In 2004, a project was carried out in collaboration between a French team (Toulouse) and a Japanese team to set up a nomenclature for Msats in the HLA region and to characterize Msats primers [3]. Recent studies of HLA Msats have furthered the understanding of the genetic organization and the extent and patterns of linkage disequilibrium (LD) within the HLA region [4].
Msats are not themselves functional, however, their inherent polymorphism and LD with HLA loci make them a robust disease-mapping tool in understanding susceptibility to autoimmune and infectious diseases [5] and in delimiting recombinations within the Major Histocompatibility Complex (MHC) [6].
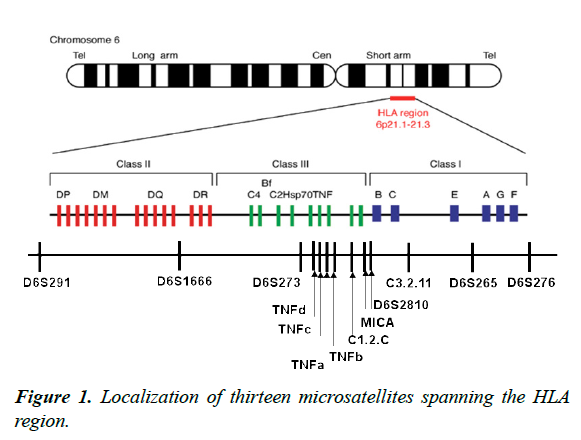
The hypothesis is that STR markers can provide useful HLA haplotype information; as such information would be of value in unrelated bone marrow transplantation. The aims of our study were to define a set of reliable HLA-region Msat (13 STRs loci were evenly distributed in the HLA region, where STRMICA, D6S2810, C3-2-11 (D6S2701), D6S265 and D6S276 were located in the HLA class I region, D6S2789 (TNFd), STR TNFc, a and b, C1_2_C (D6S2800) in the HLA III region and D6S291, D6S1666 in the HLA class II region) (Figure 1), to evaluate their LD with HLA alleles; and to test Msats ability to predict HLA typing.
Materials and Methods
The study population consisted of 123 healthy unrelated individuals originating from the South Tunisia, and typed for HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 [7]
Microsatellite selection and characteristic display
D6S291, D6S1666, D6S273, D6S2789 (TNFd), STR TNFc, a et b, C1_2_C (D6S2800), STR-MICA, D6S2810, C3-2- 11(D6S2701), D6S265 and D6S276 were selected from the dbMHC Msat resource (dbMHCMsat resource). The dbMHC Msat portal has been developed as an online database of MHC Msat markers by Gourraud et al. (available at http://www.ncbi.nlm.nih.gov/projects/gv/mhc/main.fcgi?cmd=init) [8]. Searching for Msats markers in dbMHC returns the following information: (i) the locus name and a graphic display of the location of each marker, (ii) a list of the primer pairs and the primer pair names, (iii) repeated motif and allele size range and (iv) a table that displayed physical mapping information. The Msats data obtained for the selected markers are summarized in Table 1.
| STR | Localization | Motif | Primer sequences (forward and reverse) (5’–3’) |
Labeling | Expected product size (bp) |
|---|---|---|---|---|---|
| D6S291 | Tg (3200kb) HLA-DPB1 |
CA | CTCAGAGGATGCCATGTCTAAAATA GGGGATGACGAATTATTCACTAACT |
6-FAM- | 196-212 |
| D6S1666=DQCAR II | Tg (2kb) HLA-DQA1 |
GT | TGATTCATAAGGCAAGAATCCAGCATATTG GCAATATCATTAAATTTGCTTTCCACAGTAT |
6-FAM- | 183-217 |
| D6S273 | Tg (205 kb) MICB |
CA | GCAACTTTTCTGTCAATCCA ACCAAACTTCAAATTTTCGG |
6-FAM- | 120-134 |
| D6S276 | Tg (6500kb) HLA-A |
CA | TCAATCAAATCATCCCCAGAAG GGGTGCAACTTGTTCCTCCT |
VIC | 194-222 |
| TNFa | Tg (57kb) MICB |
AC | GCCTCTAGATTTCATCCAGCCA CCTCTCCCCCTGCAACACACA |
VIC | 98-122 |
| TNFb | Tg (3.5kb) TNF B |
TC | GCACTCCAGCCTAGGCCACAGA tgtgtgttgcaggggagagagg | VIC | 116-130 |
| TNFd= D6S2789 |
Tg (77kb) MICB |
AG | TCATTCCAGCTATCGCAAGG AGATCCTTCCCTGTGAGTTC |
VIC | 193-207 |
| TNFc | Tg (61kb) MICB |
TC | gggaggtctgtcttccgccg cgttcaggtggtgtcatggg |
PET | 97-99 |
| D6S265 | Tg (106kb) HLA-A |
CA | ACGTTCGTACCCATTAACCT ATCGAGGTAAACAGCAGAAA |
NED | 116-138 |
| MICA-STR | Tg (55kb) HLA-B |
GCT | CCTTTTTTTCAGGGAAAGTGC CCTTACCATCTCCAGAAACTGC |
NED | 181-193 |
| D6S2810 | Tg (24kb) HLA-B |
GT | CTACCATGACCCCCTTCCCC CGTACCACAGTCTCTATCAGTCCAG |
NED | 328-360 |
| C1_2_C= D6S2800 |
Tg (85kb) HLA-B |
AC | GGATCCTAGGAACTCCCTCCTG GAGCAGAAGGGAGATGAAATGG |
NED | 234-262 |
| C3-2-11= D6S2701 |
Tg (494kb) HLA-A |
GA | AGATGGCATTTGGAGAGTGCAG TCCTTACAGCAGAGATATGTGG |
NED | 183-225 |
Table 1. Characteristics of the 13 short tandem repeat.
DNA samples, PCR amplification and genotyping
All samples were electrophoresed on the ABI Prism_310 sequencer and analyzed using GENESCANv3.1.2. Allele assignment was based on the amplicon size (the number of base pairs: bp).
Statistical methods
STR characteristics study Allele frequencies, expected numbers of genotypes, homozygotes, heterozygotes and PIC (polymorphism information content) as well as the Hardy– Weinberg equilibrium (HWE) were calculated using the Power Marker program. Linkage disequilibrium study Haplotype frequencies and LD were computed using the PyPop program. Patterns of overall LD were measured as Wn and D’. Both Wn and D’are standardized measures that range from zero to one, with higher values indicating stronger LD. While the two statistics are correlated, they are influenced differently by various aspects of the strength of LD, such as sensitivity to the number of alleles or estimation of low-frequency haplotypes.
Haplotype prediction by Msats study, we introduced a measure of STR diversity of specific HLA-A-B-DRB1 defined haplotypes called ‘haplotype specific heterozygosity’ (HSH). The HSH is the heterozygosity of a particular STR given a specific HLA haplotype. [9]. It is computed separately for each HLA haplotype by normalizing the STR allele frequencies found on the specific HLA haplotype and then calculating the above heterozygosity statistic using the normalized frequencies.
The normalized frequencies for these haplotype specific STR
alleles are 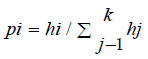 and then
and then 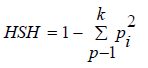 where k
is the number of STR alleles observed on the specific HLA-AB-
DRB1 haplotype, and h1, . . . hk are the frequencies of the
four-locus STR–HLA-A-B-DRB1 haplotypes.
where k
is the number of STR alleles observed on the specific HLA-AB-
DRB1 haplotype, and h1, . . . hk are the frequencies of the
four-locus STR–HLA-A-B-DRB1 haplotypes.
For the prediction of specific HLA haplotypes by STR alleles, frequencies for haplotypes consisting of HLA-A, HLA-B, HLA-DRB1, and one or more Msats were estimated.
The sensitivity is defined as the probability of observing the Msat allele(s) given a particular HLA haplotype. The specificity is the probability of not observing the Msats allele(s) given that the HLA haplotype was not observed. The positive predictive value (PPV) is defined as the probability of observing the HLA haplotype given that the specific Msat allele(s) was observed. The negative predictive value is the probability of not observing the HLA haplotype given that the specific Msats allele(s) was not observed. Higher values for each of these statistics indicate, in slightly different ways, that there is a strong association of the Msats allele(s) with the HLA haplotype.
Results
Characteristics of the 13 STR
The highest level of polymorphism, measured by the number of alleles, was observed for C3-2-11 (21 alleles) and the lowest with the TNFc marker (tow alleles) (Table 2). At each Msats locus, one to four major alleles represented 50% of the total frequency. HWE was verified for all STRs (p non-significant), except the TNFb and D6S1666 Msats. The percentage of individuals homozygous for these two Msats was 59% for D6S1666 and 43% for TNFb.
| STR | MFA (%) | Number of alleles | Heterozygosity | PIC | HWE (p) |
|---|---|---|---|---|---|
| D6S276 | 29.2 | 13 | 0.91 | 0.84 | 0.778 |
| HLA-A | 21.5 | 16 | 0.80 | 0.88 | 0.333 |
| D6S265 | 35.5 | 10 | 0.79 | 0.78 | 0.857 |
| C3-2-11 | 14.6 | 21 | 0.93 | 0.87 | 0.800 |
| HLA-C | 21.1 | 13 | 0.80 | 0.87 | 0.269 |
| HLA-B | 11.7 | 26 | 0.91 | 0.94 | 0.851 |
| D6S2810 | 17.4 | 15 | 0.86 | 0.88 | 0.093 |
| STR-MICA | 44.3 | 6 | 0.69 | 0.65 | 0.937 |
| C1_2_C | 25.6 | 14 | 0.86 | 0.80 | 0.439 |
| TNFb | 41.8 | 8 | 0.57 | 0.70 | 8 × 10-5 |
| TNFa | 18.6 | 13 | 0.82 | 0.87 | 0.400 |
| TNFc | 69.6 | 2 | 0.44 | 0.33 | 0.659 |
| D6S2789 | 43.0 | 8 | 0.67 | 0.66 | 0.968 |
| D6S273 | 28.5 | 7 | 0.85 | 0.79 | 0.504 |
| HLA-DRB1 | 16.2 | 13 | 0.90 | 0.87 | 0.483 |
| D6S1666 | 24.3 | 14 | 0.41 | 0.84 | 1 × 10-26 |
| HLA-DQB1 | 31.3 | 7 | 0.78 | 0.77 | 0.150 |
| D6S291 | 23.1 | 10 | 0.82 | 0.80 | 0.961 |
Table 2. Polymorphism of the 13 short tandem repeats.
LD between Msats and HLA
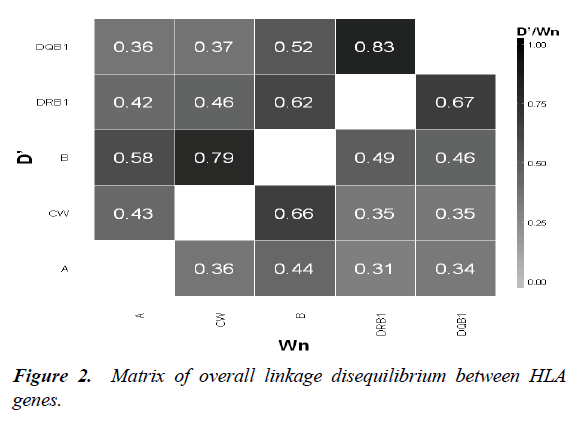
We measured LD between HLA loci. The strongest LD was observed between HLA-DRB1-DQB1 (Wn = 0.67 ; D’= 0.83) and HLA-C-B (Wn=0.66 ; D’= 0.79) (Figure 2). Regarding HLA-Msats LD, Msat had the highest degree of LD with the nearest HLA locus. The strongest associations were observed between HLA-A-D6S265 (Wn = 0.55; D’ = 0.76), HLA-BD6S2810 (Wn=0.61; D’=0.8), HLA-B-STR-MICA (Wn=0.7 ; D’=0.78) and HLA-DR-D6S1666 (Wn=0.45 ; D’=0.67). Each of these associations was significant
HSH
The HSH provides a summary of the distribution of Msats alleles of HLA-defined haplotypes and gives additional haplotype specific information about the diversity of the STR and how this diversity varies from one HLA-defined haplotype to another. Diversity of Msats among the six most frequent haplotypes in our group is displayed in Table 3. This table contains the overall heterozygosity (gene diversity) index and HLA-A-B-DRB1 HSH for each STR marker.
| HLA Haplotypes | |||||||
|---|---|---|---|---|---|---|---|
| Marqueur | Het | A1-B8-DR3 (7) | A2-B50-DR7 (6) | A1-B52-DR15 (5) | A1-B58-DR7 (4) | A23-B44-DR7 (4) | A24-B35-DR4 (4) |
| D6S276 | 0.91 | HSH=0.24 | HSH=0.48 | HSH=0.66 | HSH=0.56 | HSH=0.62 | HSH=0.62 |
| HLA-A | |||||||
| D6S265 | 0.79 | HSH=0 (a10) | HSH=0 (a10) | HSH=0 (a10) | HSH=0 (a10) | HSH=0 (a10) | HSH=0.37 |
| C3-2-11 | 0.93 | HSH=0.24 | HSH=0.27 | HSH=0.5 | HSH=0.5 | HSH=0.44 | HSH=0 (a23) |
| HLA-C | -- | -- | -- | -- | -- | ||
| HLA-B | |||||||
| D6S2810 | 0.86 | HSH=0.24 | HSH=0.44 | HSH=0 (a7) | HSH=0 (a16) | HSH=0 (a12) | HSH=0.37 |
| MICA | 0.69 | HSH=0.24 | HSH=0 (a6) | HSH=0 (a6) | HSH=0 (a9) | HSH=0 (a6) | HSH=0.37 |
| C1_2_C | 0.86 | HSH=0 (a18) | HSH=0.32 | HSH=0.48 | HSH=0 (a17) | HSH=0 (a15) | HSH=0.44 |
| TNFb | 0.57 | HSH=0.40 | HSH=0.27 | HSH=0.32 | HSH=0 (b5) | HSH=0.62 | HSH=0.37 |
| TNFa | 0.82 | HSH=0.24 | HSH=0.32 | HSH=0.32 | HSH=0.65 | HSH=0.44 | HSH=0 (a2) |
| TNFc | 0.44 | HSH=0 (c1) | HSH=0 (c1) | HSH=0 (c1) | HSH=0 (c1) | HSH=0.37 | HSH=0.37 |
| D6S2789 | 0.67 | HSH=0.24 | HSH=0.27 | HSH=0 (a16) | HSH=0.37 | HSH=0 (a14) | HSH=0.44 |
| D6S273 | 0.85 | HSH=0.44 | HSH=0.27 | HSH=0.56 | HSH=0.5 | HSH=0.37 | HSH=0 (a15) |
| HLA-DRB1 | |||||||
| D6S1666 | 0.41 | HSH=0.24 | HSH=0.61 | HSH=0.32 | HSH=0 (a11) | HSH=0.37 | HSH=0.37 |
| HLA-DQ | |||||||
| D6S291 | 0.82 | HSH=0.62 | HSH=0.37 | HSH=0.48 | HSH=0 (a9) | HSH=0 (a9) | HSH=0.44 |
Table 3. Marker heterozygosity and haplotype specific heterozygosity in South Tunisian population.
Each of the six HLA-A-B-DR haplotypes described in Table 3 had at least one Msat with an HSH value of zero indicating that only one Msat allele was observed at that Msat locus for the specific HLA haplotype. HSH was zero for the following Msat–HLA haplotypes: C3-2-11 a23 allele and HLA-A24- B35-DR4 haplotype, D6S2810 a7 allele and HLA-A1-B52- DR15 haplotype, STR-MICA a9 allele and HLA-A1-B52- DR15 haplotype, C1_2_Ca15 allele and HLA-A23-B44-DR7 haplotype and finally a16 of the D6S2789 with HLA-A1-B52- DR15. D6S276, the farthest Msat from HLA-A locus, had no allele with a null HSH.
Prediction of HLA-A-B-DRB1 haplotype by Msats
We assessed the utility of Msats to predict specific HLA-AB- DRB1 haplotypes. We focused on the three most common HLA haplotypes: “HLA-A1-B8-DR3, HLA-A2-B50-DR7 and HLA-A1-B52-DR15”, which had frequencies of 3%; 2.24% and 2.2%, respective. We chose two sets of three markers with the highest LD and lowest HSH. D6S265 (a10) along with C1_2_C (a18) alleles, were selected for A1-B8-DR3 haplotype. D6S265 (a10) and STR-MICA (a6) were chosen to predict the HLA-A2-B50-DR7 haplotype. Regarding the HLA-A1-B52- DR15 haplotype, combination of the alleles markers D6S265 (a10), D6S2810 (a7), STR-MICA (a6) and D6S2789 (a16) was necessary for haplotype prediction.
Finally the ability of markers to predict HLA-A1-B8-DR3 haplotypes, HLA-A2-B50-DR7 and HLA-A1-B52-DR15 were evaluated by the study of sensitivity, specificity, positive predictive value and negative predictive value presented in Table 4. Despite a sensitivity of 100% for D6S265 (a10) with HLA-A1-B8-DR3 and (a18) of C1_2_C with HLA-A1-B8- DR3, the alleles PPV, considered separately, were low.
| Haplotype A-B-DR | STR | STR Allele | Sensitivitya | Specificityb | PPVc | NPVd | Number |
|---|---|---|---|---|---|---|---|
| A1-B8-DR3 | D6S265 | a10 | 1 | 0.67 | 0.08 | 1 | 7 |
| A1-B8-DR3 | C1_2_C | a18 | 1 | 0.85 | 0.16 | 1 | 7 |
| A1-B8-DR3 | D6S265- C1_2_C | a10-a18 | 1 | 0.97 | 0.50 | 1 | 7 |
| A2-B50-DR7 | D6S265 | a10 | 1 | 0.67 | 0.07 | 1 | 6 |
| A2-B50-DR7 | STR-MICA | A6 | 1 | 0.57 | 0.05 | 1 | 6 |
| A2-B50-DR7 | D6S265- STR-MICA | a10-a6 | 1 | 0.86 | 0.15 | 1 | 6 |
| A1-B52-DR15 | D6S265 | a10 | 1 | 0.67 | 0.06 | 1 | 5 |
| A1-B52-DR15 | D6S2810 | a7 | 1 | 0.85 | 0.12 | 1 | 5 |
| A1-B52-DR15 | STR-MICA | a6 | 1 | 0.57 | 0.05 | 1 | 5 |
| A1-B52-DR15 | D6S2789 | a16 | 0.8 | 0.9 | 015 | 0.99 | 5 |
| A1-B52-DR15 | D6S265- D6S2810- STR-MICA- D6S2789 | a10-a7-a6-a16 | 0.8 | 1 | 1 | 0.99 | 5 |
b: Specificity; The probability of not observing the particular Msat allele given the absence of the particular HLA haplotype.
c:Positive predictive value; The probability of observing the particular HLA haplotype given the presence of the particular Msat allele.
d: Negative predictive value; The probability of not observing the particular HLA haplotype given the absence of the particular Msat allele.
e: Number; The number of haplotypes.
Table 4. Prediction of HLA haplotypes by Short Tandem Repeat in South Tunisian population.
The combination of the two markers D6S265 (a10) and (a18) of C1_2_C was necessary to have a higher PPV (0.5) and consequently to predict the haplotype HLA-A1-B8-DR3. In our study, we found only two PPVs with values greater than 50%: HLA-A1-B8-DR3, D6S265 (a10) -C1_2_C (a18) and HLA-A1- B52-DR15, D6S265 (a10) - D6S2810 (a7) - STR-MICA (a6) - D6S2789 (a16).
Discussion
In the current study, we investigated 13 STR markers located around or within the HLA-A, B, C, DR and DQ loci.
All markers were found to respond to HWE in our Southern Tunisian population except D6S1666 and TNFb. Deviation from HWE for the two latter Msats could be indicative of a mis identification of unamplified alleles. Msats respecting The HWE can be used in population studies and in Msat-disease association analysis.
In addition to the strong LD reported between the different HLA loci (B-C and DRB1-DQB1), we also found that the Msat markers nearest to the HLA loci were those with the strongest LD: HLA-A-D6S265, HLA-B-D6S2810, HLA-B-STR-MICA and HLA-DR-D6S1666.
Comparing our results with a study of Caucasian haematopoietic stem cell donors [10]. We found that some of Msats have the same strong DL: HLA -A-D6S265 and HLA-B-D6S2810. Regarding statistical parameters, we used LD and HSH analysis to ascertain the best MSAts for HLA prediction. A marker in strong LD with an HLA locus and with a low value of HSH is the most appropriate for predicting HLA alleles.
Markers in moderate or weak LD with HLA loci or with higher HSH will not be useful in screening for common HLA haplotypes. On the other hand, these markers may be useful for identifying regions in the MHC that may be relevant for donor matching in transplantation and in investigating potential disease susceptibility genes in addition to known HLA effects. For prediction Accuracy we found that PPV, the probability of observing the particular HLA haplotype in the presence of a particular Msats allele, was the most relevant statistical parameters. H-S [11] have established a three-microsatellite (D6S2666, D6S2665, D6S2446) haplotyping method that can serve as a surrogate for DRB1 genotyping with very good sensitivity and specificity for most of the major DRB1 alleles in Caucasian and Asian groups. A high degree of haplotype conservation was demonstrated in healthy controls as well as rheumatoid arthritis patients.
Besides Msats [12] employed SNP markers for Predicting Classical HLA Alleles in African, Asian and Caucasian groups. They defined a panel of ~100 SNPs typed across the HLA region for predicting both rare and common HLA alleles up to 95% accuracy. In their study, haplotype phase of trio data was used for reconstructing the HLA-SNP haplotypes. Underestimation of heterozygosity was the most important imitation of SNPbased method [13,14].
Conclusion
In conclusion, to refine the prediction accuracy, it is necessary to associate different Msats and SNP markers in a large cohort. Such an approach can provide a low-cost HLA-typing method that is useful in many clinical settings.
References
- Foissac A, Salhi M, Cambon-Thomsen A, et al. Microsatellites in the HLA region: 1999 Update. Tissue Antigens. 2000;55(6);477-509.
- Gourraud PA, Mano S, Barnetche T, et al. Integration of microsatellite characteristics in the MHC region: A literature and sequence based analysis. Tissue Antigens. 2004;543-55.
- Gourraud PA, Feolo M, Hoffman D, et al. The db MHC microsatellite portal: a public resource for the storage and display of MHC microsatellite information. Tissue Antigens. 2006:67:395-401.
- Gourraud PA, Utilisation d’analyses des haplotypes dans l'organisation de la transplantation de cellules souches hématopoeitiques. Toulouse III 2005.
- Lee HS, Li A, Rodine P, et al. Microsatellite typing for DRB1 alleles: Application to the analysis of HLA associations with rheumatoid arthritis. Genes and Immunity. 2006;533–543.
- Lahiani NM, Kamoun A, Bellaaj H, et al. Molecular analysis of crossing-over in the CMH in two Tunisian families with aplastic bone marrow. Pathologie Biologie. 2009; 5:383-387.
- Lancaster A, Nelson MP, Meyer D, et al. PyPop: A software framework for population genomics: analyzing large-scale multi-locus genotype data. Pacific Symp Biocomput. 2003 ; 514–25.
- Lancaster A. Haplotype Estimation and Linkage Disequilibrium Methods: Manual: 2011; Version 0.1.8
- Lander E, Linton L, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001.
- Mahfoudh N, Ayadi I, Kamoun A, et al. Analysis of HLA-A,-B,-C,-DR,-DQ polymorphisms in the South Tunisian population and a comparison with other populations. Annals of Human Biology. 2013;41-47.
- Malkki M, Gooley T, Horowitz M, et al. Mapping MHC-Resident Transplantation Determinants. Biology Blood and Marrow Transplantation. 13(8):2007; 986–995.
- Malkki M, Single R, Carrington M, et al. MHC microsatellite diversity and linkage disequilibrium among common HLA-A, HLA-B, DRB1 haplotypes: Implications for unrelated donor hematopoietic transplantation and disease association studies. Tissue Antigens. 66(2): 2005;114–24.
- Subramanian S, Mishra RK, Singh L, et al. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 2003;4(2):R13.
- Leslie S, Donnelly P, McVean G, et al. A statistical method for predicting classical HLA alleles from SNP Data. The American Journal of Human Genetics. 2008; 82;48–56.