Review Article - Current Pediatric Research (2021) Volume 25, Issue 11
The variant VNTR INS -23/Hph1 A>T (rs689) in the insulin gene has been related with TIDM in children.
Asmaa Lateef Neama1, Mazin Majid Mohammed Ali2, Alaa Jumaah Manji Nasrawi3*
1Department of Pediatrics Al-Muthanna, Al-Muthanna, Iraq
2Department of Pediatrics, Al Muthanna Health Directorate, Samawah, Iraq
3Department of Pediatrics, Medical College, Kufa University, Kufa, Iraq
- Corresponding Author:
- Alaa Jumaah Manji Nasrawi
Department of Pediatrics
Medical College
Kufa University
Kufa
Iraq
Accepted date: November 23th, 2021
Abstract
T1DM is a complicated condition influenced with both inherited and biological conditions. Many studies have found a link between genetic variations and disease; however, the significance of specific gene loci is still unknown. The objectives of this paper were to see if there was a link between both the rs689 gene polymorphism and type1 diabetes in children. The A>T polymorphism in the INS-23/Hph1 gene is a significant risk susceptibility locus for TIDM in children, with T allele carriers having a 2.56 fold greater risk of getting the disease. Some of the conclusions are consistent with previous research, while others are not. The discrepancy in results could indicate that many mechanisms are at play, with central tolerance vs. peripheral tolerance being the most relevant aspect. This highlights the importance of doing susceptibility research on a local scale; results from one ethnic group or population cannot be applied to others. The study has other disadvantages, such as a small number of patients and a matched control group.
Keywords
TIDM, Insulin gene, PCR, RFLP, Gen polymorphism.
Introduction
Diabetes is defined as a series of metabolic diseases that lead to the development of Diabetes Mellitus (DM) it is caused by a deficiency in insulin secretion, insulin efficiency, or both [1]. In type 1 diabetes, hyperglycemia arises when the pancreas fails to produce enough insulin or cells due to beta-cell insufficiency. That chemical effectively symbolizes non-diabetes, type 2 diabetes. Kind 1 diabetes is a type of diabetes that is Not Insulin-Dependent (NIDDM). One of the most deadly inherited autoimmune illnesses; it affects children in their youth and early adolescence. The loss of beta cells causes people with this kind to stop releasing the hormone insulin [2]. The basic element for T1DM pathogenesis is thought to be a mix of genetic susceptibility and environmental factors [3]. Alleles or genetic variants associated with T1DM provide vulnerability to or prevention from the syndrome depending on the environment [3]. HLA genes are the most important in determining TIDM vulnerability [4]. Outside of the Human Leukocyte Antigen (HLA) area, T1DM has been associated to more than 50 loci [5]. The genetic makeup of an individual, which includes a balance of susceptible and protective genes, determines the age at which T1DM develops. Genetic variants in the noncoding area of the Insulin (INS) gene (IDDM2) are second only to HLA in determining susceptibility to or prevention from T1DM [6]. At this position, a Variation Nucleotide Tandem Repeat (VNTR) mini-satellite can be found at the 5 terminus of the insulin gene [6]. VNTR size is divided into three classes: class I (26-63 repeats), class II (approximately 80 repeats), and class III (140–200 repeats). Insulin variants are linked to the VNTR region, so they can be used as markers for these gene variants [7]. The 223HphI A>T SNP, a differential sequence in the INS gene, has just been found to be in high genetic linkage with the VNTR alleles, with the A allele related to the small (class I) VNTR allele and the T allele associated to the long (class III) VNTR allele [8]. The contribution of the HLA region and CTLA-4 has been the topic of study on the genetic basis of T1DM undertaken in AL-Muthanna, Iraq, as well as those conducted by the research team [8–10]. Just one previous study looked at the role of the INS gene in sensitivity to TIDM risk in a group of 25 participants. In the present study, the INS-23/Hph1 A>T (rs689) was investigated in diabetes children, non-diabetic siblings and parents, and also healthy controls. This, among other genetic data, could help predict whether a child born into a diabetic family will develop the disease, enabling for earlier treatment and improved care of diabetic youngsters. In Iraq, there are limited genetic investigations, particularly in the area of phenotype variation of genes that cause hereditary illnesses. For this study, the ARFLP technique was used to analyze the polymorphism of the genotype of individual using special primers and through which the allele is determined with PCR calculation) is the Cytokines whose repeats in the gene for samples studied [9].
Materials and Methods
Fifty blood samples were collected from children with diabetes-type 1 and 50 samples from healthy children (the standard group) from central children's hospitals and diabetes center in Al-Muthanna governorate Iraq for the period from February 2021 until April of the same year. The children's ages ranged from 6-13 years old. Blood was collected in 5 ml blood collection tubes containing EDTA as an anticoagulant, which was used for DNA extraction. The samples were diagnosed by specialists in the diabetes unit at children's central hospital and the diabetes center. EasyPure® blood genomic DNA Kit used. The primers are prepared depending on the manufacturing instruction by dissolving the lyophilized primers with TE (Tris-EDTA) buffer to make stock solution of concentration of 100 pmole/Ml, On spinning down and stay overnight at 4°C , each primer's work solution was made by reducing the standard solution with TE buffer to reach final working solution (10 pmole/Ml). The primers used in this study show in Table 1.
| Primer | sequences | References |
|---|---|---|
| MPB | 5′-FAGCAGG TCT GTT CCA AGG-3′ R 5'AGCATCGAGTCGATCGCGGA '3 | 5 |
Table 1. Show primer and sequences used.
PCR master mix reaction was prepared by using (one taq quick-load) PCR Kit and this master mix done according to company instructions as shown. The PCR setup steps show in Table 2.
| Component | 25 μl Reaction |
|---|---|
| 10 µM Forward Primer | 0.5 µl |
| 10 µM Reverse Primer | 0.5 µl |
| Template DNA | 1.5 µl |
| OneTaq Quick-Load 2X Master Mix with Standard Buffer | 12.5 μl |
| Nuclease-free water | 10 μl |
| Total | 25 μl |
Table 2. The PCR setup steps.
Thermocycling conditions for a routine PCR show in Table 3.
| Steps | Temperatures | Time |
|---|---|---|
| Initial Denaturation | 94°C | 30 seconds |
| 30 Cycles | 94°C | 30 seconds |
| 55°C | 60 seconds | |
| 68°C | 1 minute | |
| Final Extension | 68°C | 5 minutes |
| Hold | 4°C-10°C | ∞ |
Table 3. Show thermo cycling conditions.
Genotyping of the INS-23/Hph1 A>T polymorphism PCR products from each sample was digested by 10000U of Hph I restriction. At 37°C for 18 hours, the amplicons were processed with Hph I restrict enzyme. Products were electrophoresed on a 4 percent agarose gel then stained with ethidium bromide after digesting. The T and A alleles were separated by 231 plus 129 bp and 191 plus 129 bp bands, accordingly. Enzyme by using the protocol (Table 4).
| Polymorphism | Restriction enzyme | PCR product (bp) | RFLP product (bp) | Mixture of restriction analyses | ||||
|---|---|---|---|---|---|---|---|---|
| Enzyme | Buffer | ddH2O | PCR product | Total volume | ||||
| INS -23/Hph1 A>T | 10,000 units/ml (Hph I) | 360 | 23,11,91,129 | 1 µl | 5 µl | 9 µl | 10 µl | 25 µl |
Table 4. Show details of restriction enzyme.
Results
DNA extraction band from blood sample show in the flowing picture of gel electrophoresis (Figure 1 and Figure 2).
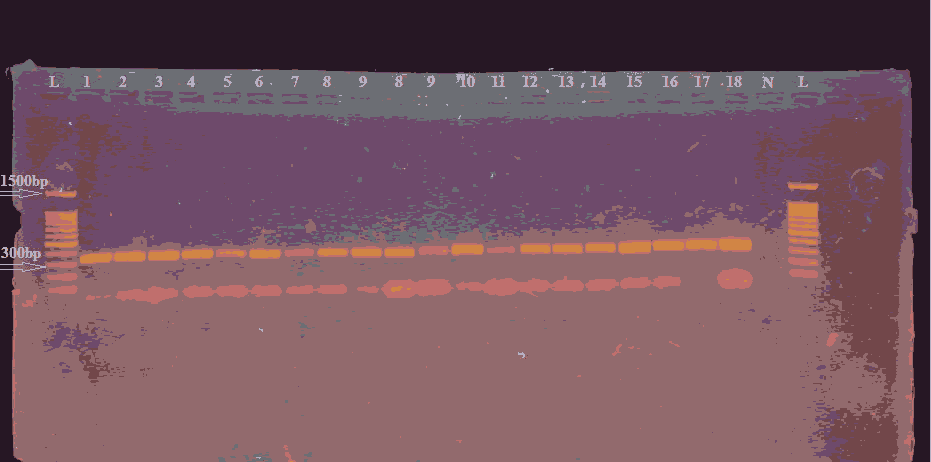
Figure 2. Gel electrophoresis for PCR product of (INS -23 primer) which show 360 bp primer TM at (55C), (Agarose 1.5% 15 min. At 100 voltage and then lowered to 70 volts, 60 min.). Visualized under UV light after staining with ethidium bromide. Lane L: DNA ladder (1500-100) bp, Lanes (1-18zW) represented positive results, lane N represent negative control.
In Table 1 shows the frequency of rs689 in the various groupings. The genotype frequency of the child's parents and non-diabetic siblings was comparable (p=0.97). In comparison to controls A was over-represented in the three family units, but TT and AT were over-represented in the controls (p=0.0014 and 0.0028, respectively) (Figure 3).
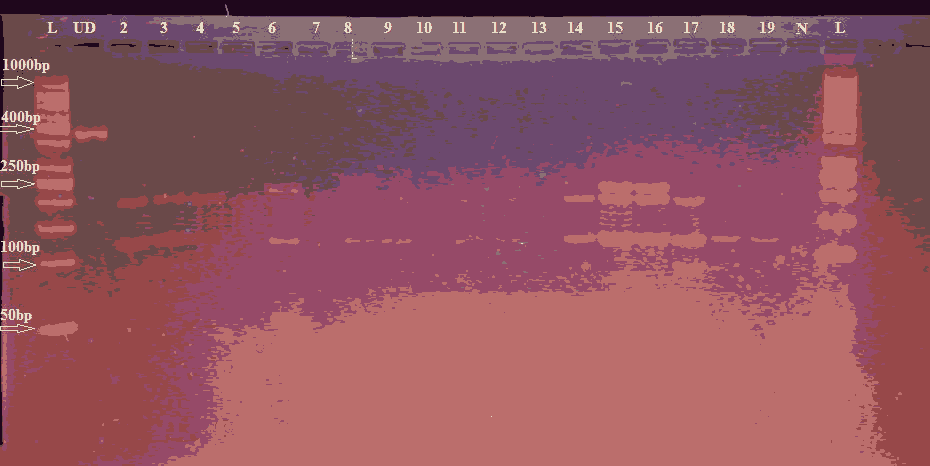
Figure 3. Agarose gels showing the enzyme-digested patterns of PCR products of INS-23/Hph1 A>T polymorphism. Electropherogram of ethidium bromide stained 4% agarose gel showing genotype pattern obtained with HphI restriction digest. Lane L: DNA ladder (50-1000) bp Lane UD refers to the undigested PCR-amplified (360 bp) fragment. Genotypes are characterized as AA (191 bp, 129 bp, and 40 bp) 40 bp fragments cannot be seen represented with (17-19) samples, AT (231 bp, 191 bp and 129 bp) represented with (15 and 16) samples, and TT (231 bp and 129 bp) represented with (6 and 8) samples. On the gel Lane (13) showed negative results, lane N represent negative control
Genotype and allele frequency shown in (Table 5).
| Group | No | INS -23/ A>T genotype | Mixture of restriction analyses | |||||
|---|---|---|---|---|---|---|---|---|
| enzyme | buffer | ddH2O | PCR product | Total volume | ||||
| INS-23/Hph1 A>T | 10,000 units/ml (Hph I) | 360 | 23,11,91,129 | 1 µl | 5 µl | 9 µl | 10 µl | 25 µl |
Table 5. Genotype and allele frequency of INS -23 A>T in type 1 diabetes mellitus and control.
The AA allele was inherited from the mother in 34 patients and from the parent in 20 patients, the AT allele was received from the parent in 23 instances and from the parent in 15 patients, and the TT allele was inherited from the parent in one incidence each (Table 6).
| Groups | INS -23/ A>T genotype | Allele frequency | |||
|---|---|---|---|---|---|
| AA | AT | TT | A | T | |
| Type 1 diabetes mellitus | 27 | 21 | 2 | 0.75 | 0.25 |
| Control | 41 | 8 | 1 | 0.1 | 0.9 |
Table 6. Genotype and allele frequency of INS -23 A>T in type 1 diabetes mellitus and control according to Hardy Weinberg equilibrium for genotype frequency the results of our experiment as fallowing.
In case to type 1 diabetes mellitus group allele frequency=0.999 which is not equal to a Hardy-Weinberg equilibrium =1. SO this population is not is nearly equal equal to a Hardy-Weinberg equilibrium based on the observations of above genotype frequencies.
Discussion
In the present study, the 223HphI A>T was evaluated in 91 diabetic children and their parents, as well as 96 healthy controls. Parents and siblings would be included since they share genotype(s) with the patients, at partly; the parents and siblings are not diabetic at this time. As according our data, T allele carriers are 2.56 times more likely to acquire TIDM [11]. The INSrs689 T variant was shown to be associated with a 2.44-fold higher risk, which is consistent with our findings. In other studies, homozygous TT genotype was linked to a 4.5-fold risk and 4.02, 4.4, and 6.96-fold hazards in three Israeli Jewish populations [12-14]. In addition to MHC (IDDMI), more than 50 Non-HLA genes have been linked to TIDM vulnerability [4]. The most efficient of them is the INS gene (IDDM2), which also plays a role in the evolution of other endocrine diseases [15].
Insulin variants have been used as a surrogate measure for gene changes in a number of studies [16-20]. Insulin variants are in linkage disequilibrium with the VNTR region. The homozygous genotypes AA and TA are linked to the risk and progression of type 1 diabetes. The type 1 genotype can be used as a risk indicator; results may correlate with immunologic findings of the TA genotype in serum of type 1 diabetes patients. IFN-γ levels are high and significant with a risk of developing IFN-γ compared to the standard sample, and in general genetic and immunological shows how important the IFN-γ gene [21-24]. It is less than homozygous for TT with type 1 disease, and the results also show the genotype Type I, ideas indicate the importance of the genetic model that was shown as a genotype associated with the risk of developing the disease. Type of genotype the results refer to the paragraphs of the studies and progress with them, as they agreed with the results of both [25-30]. In another study of the IFN-γ gene by blocking the IFN-γ receptor gene in Iranian patients with type 1 diabetes, the gene site has a negative association with the disease, and this site is considered as a protective indicator in the resistance of diabetes mellitus-type 1 in people with diabetes. The first type has the activity of IFN-γ.
The A T1DM-risk allele, which was related to lower insulin translation stages in the thymus and ineffective deletion of insulin-specific auto-reactive T cells in other studies, has been proposed as a mechanism for the association [31-33]. Several aspects of this explanation are debatable. To begin with, the genotype associated with lower insulin production in the thymus is simultaneously associated with increased insulin synthesis in the pancreas. This brings us to the second point to consider: In autoimmune diseases, central tolerance isn't the only mechanism at action; peripheral intolerance plays a role as well [33].
Conclusion
However, other studies have shown that overexpression of IFN-γ for the purpose of degrading Macrophage cells activates macrophage IFN-γ and acts as IL-β-cell toxicity. Besides this mechanical explanation, ethnic differences, such as the comparative genotype frequency in various groups, which changes from one group to the next, have a significant impact on risk susceptibility associated with gene polymorphisms.
References
- Craig M, Hattersley A, Donaghue K. Definition, epidemiology and classification of diabetes in children and adolescents. Pediatr Diabetes 2009; 10(12): 3-12.
- American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabet Care 2014; 37(1): S81-S90.
- Kantarova D, Buc M. Genetic susceptibility to type 1 diabetes mellitus in humans. Physiol Res 2007; 56(3): 255-66.
- Ehehalt S, Dietz K, Willasch AM, et al. Prediction model for the incidence and prevalence of type 1 diabetes in childhood and adolescence: evidence for a cohort-dependent increase within the next two decades in Germany. Pediatr Diabet 2012; 13(1): 15-20.
- Atkinson MA, Eisenbarth GS. Type 1 diabetes: new perspectives on disease pathogenesis and treatment. Lancet 2001; 358(9277): 221-9.
- Rabinovitch A. Autoimmune diabetes. Sci Med 2000; 7(3): 18-27.
- Pociot F, McDermott MF. Genetics of type 1 diabetes mellitus. Genes Immun 2002; 3(5): 235-49.
- Pociot F, Akolkar B, Concannon P, et al. Genetics of type 1 diabetes: what’s next? Diabetes 2010; 59(7): 1561-71.
- Todd JA. Etiology of type 1 diabetes. Immunity 2010; 32(4): 457-67.
- Todd JA, Walker NM, Cooper JD, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet 2007; 39(7): 857-64.
- Expert Committee on the Diagnosis and Classification of Diabetes Mellitus.Report of the expert committee on the diagnosis and classification of diabetes mellitus. Diabet Care 2003; 26(1): 5-20.
- Sambrook J, Frisch EF, Maniatis T. Molecular cloning: A laboratory manual. (2nd edn) New York: Cold spring harbor laboratory press.n1989.
- Novota P, Kolostova K, Pinterova D, et al. Interleukin IL-18 gene promoter polymorphisms in adult patients with type 1 diabetes mellitus and latent autoimmune diabetes in adults. Immunol Lett 2005; 96(2): 247-51.
- Dong GP, Yu ZS, Liang L, et al. IL-18 gene promoter –137C/G and –607C/A polymorphisms in Chinese Han children with type 1 diabetes mellitus. Int J Immunogenet 2007; 34(2): 75-9.
- Mojtahedi Z, Naeimi S, Farjadian S, et al. Association of IL-18 promoter polymorphisms with predisposition to Type 1 diabetes. Diabet Med 2006; 23(3): 235-9.
- Massoud A, Bahai NS, Massoud M, et al. IL-18 gene polymorphism in type I diabetic patients: a case-control study. Tehr Univ Med J 2009; 67(1): 20-4.
- Kretowski A, Mironczuk K, Karpinska A, et al. Interleukin-18 promoter polymorphisms in type 1 diabetes. Diabetes 2002; 51(11): 3347-9.
- Urcelay E, Santiago JL, de la Calle H, et al. Interleukin-10 polymorphisms in Spanish type 1 diabetes patients. Genes Immun 2004; 5(4): 306-9.
- Reynier F, Cazalis MA, Lecoq A, et al. Lack of association of IL-10 promoter gene variants with type 1 diabetes in a French population. Hum Immunol 2006; 67(4–5): 311–7.
- Mohebbatikaljahi H, Menevse S, Yetkin I, et al. Study of interleukin-10 promoter region polymorphisms (–1082A/G, –819T/C and –592A/C) in type 1 diabetes mellitus in Turkish population. J Genet 2009; 88(2): 245-8.
- Ide A, Kawasaki E, Abiru N, et al. Genetic association between interleukin-10 gene promoter region polymorphisms and type 1 diabetes age-at-onset. Hum Immunol 2002; 63(8): 690-5.
- Yanagawa T, Maruyama T, Gomi K, et al. Lack of association between CTLA-4 gene polymorphism and IDDM in Japanese subjects. Autoimmunity 1999; 29(1): 53-6.
- Ma Y, Tang X, Chang W, et al. CTLA-4 gene A/G polymorphism associated with diabetes mellitus in Han Chinese. Chin Med J (Engl) 2002; 115(8): 1248-50.
- Nejentsev S, Cooper JD, Godfrey L, et al. Analysis of the vitamin D receptor gene sequence variants in type 1 diabetes. Diabetes 2004; 53(10): 2709-12.
- Lemos MC, Fagulha A, Coutinho E, et al. Lack of association of vitamin D receptor gene polymorphisms with susceptibility to type 1 diabetes mellitus in the Portuguese population. Hum Immunol 2008; 69(2): 134-8.
- Wang Q, Xi B, Reilly KH, et al. 2012. Quantitative assessment of the associations between four polymorphisms (FokI, ApaI, BsmI, TaqI) of vitamin D receptor gene and risk of diabetes mellitus. Mol Biol Rep 2012; 39(10): 9405-14.
- Tang S, Peng W, Wang C, et al. Association of the PTPN22 gene (+1858C/T, –1123G/C) polymorphisms with type 1 diabetes mellitus: A systematic review and meta-analysis. Diabet Res Clin Pract 2012; 97(3): 446-52.
- Xuan C, Lun LM, Zhao JX, et al. PTPN22 gene polymorphism (C1858T) is associated with susceptibility to type 1 diabetes: A meta-analysis of 19,495 cases and 25,341 controls. Ann Hum Genet 2013; 77(3): 191-203.
- Lavrikova E, Nikitin AG, Seregin Iu A, et al. Association of the C1858T polymor- phism of the PTPN22 gene with type 1 diabetes. Mol Biol 2009; 43(6): 1040–3.
- Santiago JL, Martinez A, Calle H, et al. Susceptibility to type 1 diabetes conferred by the PTPN22 C1858T polymorphism in the Spanish population. BMC Med Genet 2007; 13(8): 54.
- Cinek O, Hradsky O, Ahmedov G, et al. No independent role of the –[1]1123G>C and+2740 A>G variants in the association of PTPN22 with type 1 diabetes and juvenile idiopathic arthritis in two Caucasian populations. Diabet Res Clin Pract 2007; 76(2): 297-303.
- Liu F, Liu J, Zheng TS, et al. The –1123G>C variant of PTPN22 gene promoter is associated with latent autoimmune diabetes in adult Chinese Hans. Cell Biochem Biophys 2012; 62(2): 273-9.
- Kawasaki E, Awata T, Ikegami H, et al. Systematic search for single nucleotide polymorphisms in a lymphoid tyrosine phosphatase gene (PTPN22): association between a promoter polymorphism and type 1 diabetes in Asian populations. Am J Med Genet A 2006; 140(6): 586-93.