Research Article - Otolaryngology Online Journal (2018) Volume 8, Issue 1
The Role of Serratia Marcescens Positive Nasal Cultures in Patients with Refractory Chronic Rhinosinusitis
- *Corresponding Author:
- Stewart H. Bernard (MD)
Otolaryngology Department
University of
Florida College of Medicine, USA
Tel: 3522735199
E-mail: stewart.bernard@ent.ufl.edu
Received date: January 07, 2018; Accepted date: January 10, 2018; Published date: January 17, 2018
Abstract
Background: The role of bacteria in Chronic Rhinosinusitis (CRS) as an instigator or propagator of the disease is controversial. There has been a great deal of research into the most commonly isolated bacteria in CRS such as Staphylococcus aureus and Pseudomonas aeruginosa, there has however been little research into the less commonly isolated microbial organisms such as Serratia marcescens.
Methods: A retrospective chart review of patients who demonstrated Serratia marcescens sinus culture or DNA pyrosequencing analysis from August 2013 to December 2016 was conducted. Clinic notes, operative reports, microbiology results, and DNA pyrosequencing results were reviewed for presenting symptoms, interventions, and outcomes.
Results: 29 patients were identified with Serratia marcescens. 26/29 (89.6%) patients had undergone functional endoscopic sinus surgery (FESS) prior to their culture showing Serratia marcescens. 17 of 29 (59%) patients grew polymicrobial culture results in addition to Serratia marcescens, most commonly Pseudomonas aeurginosa (24.1%) and Staphylococcus aureus (24.1%). Combining the culture sensitivities from all 29 patients serratia had sensitivity three orally available antibiotics: levofloxacin (97%), ciprofloxacin (92%), and bactrim (97%).
Conclusions: This is the first literature to examine the characteristics of patients with Serratia Marcescens in sinus disease patients to our knowledge. Serratia marcescens infection in the sinuses may be a post surgical disease, and it appears Levofloxacin, Ciprofloxacin, and TMP/SMX are all oral anti-microbials to which Serratia marcescens is consistently susceptible. The extent to which Serratia Marcescens an instigator of recalcitrant disease or a bystander is unclear at this time and warrants further investigation.
Keywords
Microbiology; Sinus flora; DNA pyrosequencing; Endoscopic sinus surgery; Antibiotic therapy; Sinus culture; Nosocomial infections; Chronic rhinosinusitis; Microbial culture techniques; Rhinology.
Introduction
Chronic Rhinosinusitis(CRS) is a disease with a massive economical and social impact [1] effecting up to 16% of patients in the united states [2]. Although the relationship between bacteria and this burdensome disease is still poorly understood [2], bacteria does seem to play a role in the disease process. Whether bacteria instigates CRS or enters secondarily and propagates the disease is controversial. There has been a great deal of research into the most commonly isolated bacteria in CRS such as Staphylococcus aureus and Pseudomonas aeruginosa [3], there has however been little research into the less commonly isolated microbial organisms. In our clinical practice we have encountered several patients who present with sinus cultures growing the pathogenic microbe Serratia marcescens.
Serratia marcenens is a gram negative facultative anaerobe classified as a member of enterobacteriacae that has been isolated in hospital-acquired infections for several decades [4]. Known for its red pigmentation, this microbe was first described in 1819 when it was noted to be the cause of an epidemic of Italian polenta turning a blood red color. From the 1800s to the 1970s Serratia marcescens was not believed to be a pathogenic organism. This belief led Serratia marcescens, with its red pigmentation, to be used in experiments throughout the 19th and 20th century as a tracer organism to track the spread of infections [5]. Since the 1970s Serratia marcescens has now been recognized as a pathogen causing meningitis, urinary tract infections, pneumonia, bacteremia, endocarditis, peritonitis, osteomyelitis, and lung abscesses. Given its frequent occurrence in hospital acquired infections Serratia marcescens has been commonly held to be a predominantly nosocomial infection [4,5].
Several studies have been done using genomics to determine the sinus microbiome of both healthy and CRS patients. Among these studies Serratia marcescens was not listed among the common organisms isolated in healthy or CRS patients [6-10]. The goal of this study was to examine the patients from our clinical practice who presented with culture positive Serratia marcescens with respect to clinical characteristics, microbiome, and antimicrobial susceptibilities.
Methods
A retrospective chart review of patients treated by the senior author at a tertiary rhinology care institution who demonstrated Serratia marcescens on sinus culture or DNA pyrosequencing analysis of cultures obtained either in clinic or during endoscopic sinus surgery from August 2013 to December 2016 was conducted. Clinic notes, operative reports, microbiology results, and DNA pyrosequencing results were reviewed for presenting symptoms, endoscopic characteristics, interventions, and patient reported outcomes. Approval of the Institutional Review Board of the University of Florida was granted for this review (IRB# 201300621).
Routine cultures were grown and analyzed by the University of Florida Health Shands Hospital microbiology lab. To perform DNA pyrosequencing brush and swab specimens were sent to Pathogenius Laboratory (Lubbock, TX) for processing. Pathogenius is a Clinical Laboratory Improvement Amendments (CLIA) certified laboratory. DNA is extracted from specimens and 16s ribosomal DNA is amplified by polymerase chain reaction (PCR) using universal eubacterial primers.1116s rDNA amplicons were then pyrosequenced. Sequence data was processed and compared against a sequence database with validated microbial sequences at the Pathogenius Laboratory.
Results
29 patients were identified with Serratia marcescens between August 2013 and July 2016 by either routine culture or DNA pyrosequencing. Mean age at time of pathogen identification was 62.33 years (range, 26 to 83 yrs). There were 14 men and 15 women.26/29(89.6%) patients had undergone functional endoscopic sinus surgery (FESS) prior to their culture showing Serratia marcescens. 14 of 29 patients (48.3%) had 1 prior FESS and 12 of 29 patients (41.4%) had 2 or more procedures. On average, there were 1.79 procedures per patient prior to Serratia marcescens, with a range of 0 to 8 procedures.
Patients had undergone an average of 3.04 courses of oral antibiotics over the past 24 months (range 0 to 8). 13 of 29 patients (44.8%) had concomitant allergic rhinitis, 12 of 29 patients (41.4%) had concomitant asthma, and zero of the patients had and underlying immune system dysfunction (e.g. Churg-Strauss syndrome, multiple myeloma, or combined variable immune deficiency)
Symptoms at time of initial positive Serratia marcescens included: nasal discharge (55%), nasal congestion/obstruction (34%), headache/facial pressure (34%), decreased sense of smell (21%), and post-nasal drip (14%).
For the patients studied routine culture provided the diagnosis in 26 cases and DNA pyrosequencing testing provided the diagnosis in 3 cases. In total there were 11 patients who had samples sent for DNA pyrosequencing testing. Of these patients 3/11 (27.2%) grew Serratia marcescens only on pyrosequencing testing, 6/11 (54.5%) grew Serratia on both routine culture and DNA pyrosequencing testing, and 2/11 (18.2%) grew Serratia on routine culture but not on DNA pyrosequencing testing.
Combining the culture sensitivities from all 29 patient’s Serratia marcescens was found to have a 100% sensitivity to Amikacin, Cefepime, Ceftazadime, Ertapenem, Gentamicin, imipenem, and meropenem. Of the orally available antibiotic, serratia had sensitivity to levofloxacin (97%), ciprofloxacin (92%), and Bactrim (97%) Table 1.
| Name | Susceptible | Resistant | Intermediate |
|---|---|---|---|
| Amikacin | 100% | 0% | 0% |
| Amoxicillin/clavulanate | 0% | 100% | 0% |
| Ampicillin | 0% | 100% | 0% |
| Ampicillin / sulbactam | 0% | 75% | 25% |
| Cefazolin | 0% | 100% | 0% |
| Cefepime | 100% | 0% | 0% |
| Cefoxitin | 15% | 20% | 65% |
| Cefotaxime | 50% | 25% | 25% |
| Cetazidime | 100% | 0% | 0% |
| Ceftriaxone | 96% | 0% | 4% |
| Cefuroxime | 0% | 100% | 0% |
| Ciprofloxacin | 92% | 4% | 4% |
| Ertapenem | 100% | 0% | 0% |
| Gentamicin | 100% | 0% | 0% |
| Imipenem | 100% | 0% | 0% |
| Levofloxacin | 97% | 4% | 0% |
| Meropenem | 100% | 0% | 0% |
| Piperacillin/Tazobactam | 60% | 0% | 40% |
| Tetracycline | 100% | 0% | 0% |
| Ticarcillin/clavulanic | 100% | 0% | 0% |
| Tobramycin | 88% | 0% | 12% |
| Trimethoprim/sulfamethoxazole | 97% | 4% | 0% |
Table 1: Antimicrobial susceptibility of Serratia marcescens.
Out of the 18 patients treated with either levofloxacin, ciprofloxacin, or bactrim 13 (72%) reported improved symptoms at follow up. For this same group of patients, 7 had repeat cultures following completion of antibiotics. 4 (57%) of these cultures no longer showed Serratia marcescens while 3 (43%) had persistent growth of Serratia marcescens despite appropriate antimicrobial treatment with levofloxacin, ciprofloxacin, or bactrim.
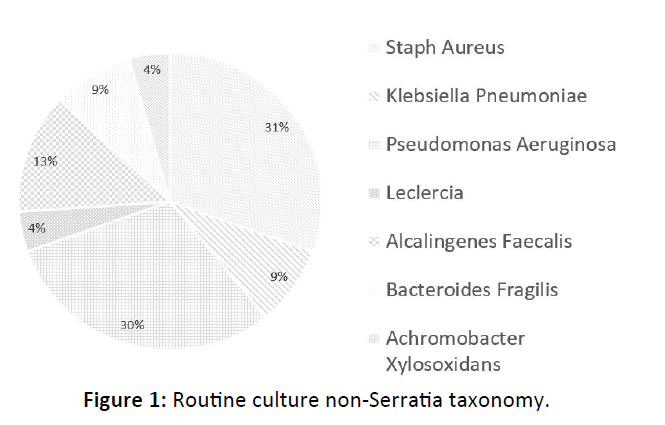
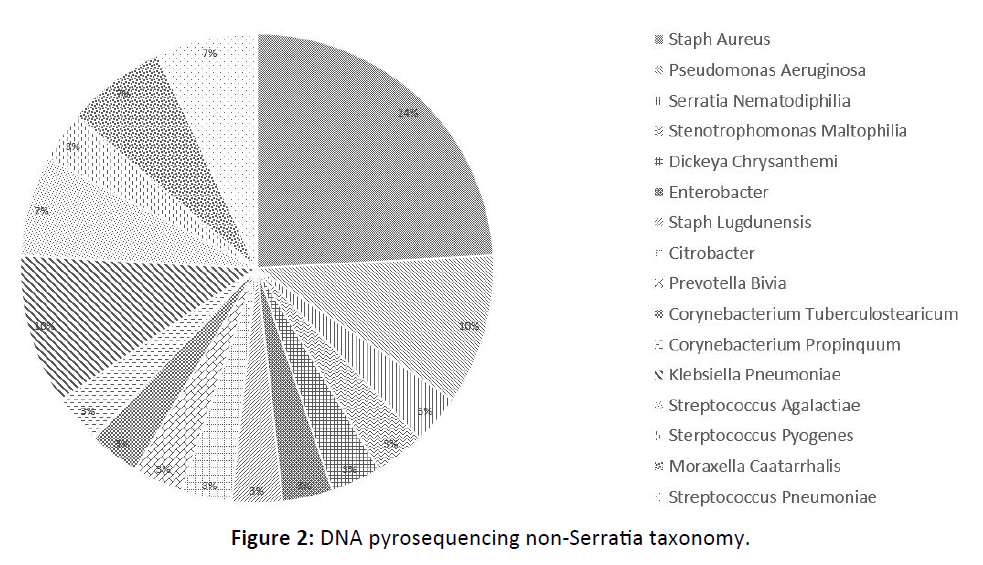
17 of 29 (59%) patients grew polymicrobial culture results in addition to Serratia marcescens, most commonly Pseudomonas aeruginosa ( 7 of 29 patients or 24.1%) and Staphylococcus aureus ( 7 of 29 patients or 24.1%). 9 of 11 (81.8%) patients with DNA analysis demonstrated non-Serratia species of bacteria, most commonly Staphylococcus aureus (7 of 11 patients or 64.6%) Figure 1, 2.
Out of the patient’s with polymicrobial infections and adequate follow up 11/15 were treated with appropriate antimicrobials (levofloxacin, ciprofloxacin, or Bactrim) with 7 of these 11 reported improvement of symptoms following antimicrobial treatment (46.67%). Of the patients without polymicrobial cultures and adequate follow up 4/9 were treated with appropriate antimicrobials with 4 patients reporting improved symptoms on follow up (44.44%). Comparing the data there was only a 2.22% difference between polymicrobial and unimicrobial patients reporting improvement from antimicrobial treatment (p=0.38) Table 2.
| Patient Id | Age | Gender | # of Previous FESS | Type of Dx | Asthma | Type of Culture | Sens to Bactrim | Sens to Cipro | Sens to Levo | Comorbid Bacteria on Routine Culture | Comorbid Bacteria on Pyro Seq |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 30 | F | 3 | CRSwNP | N | Routine only | Y | Y | Y | ||
| 2 | 53 | F | 5 | CRSwNP | Y | Routine & Pyro | Y | Y | Y | Staphylococcus aureus | Staphylococcus aureus |
| 3 | 72 | F | 4 | CRSwNP | Y | Routine & Pyro | Y | Y | Y | klebsiella and Staphylococcus aureus | Staphylococcus aureus |
| 4 | 69 | F | 1 | CRSsNP | N | Routine & Pyro | N | 2 | Y | Pseudomonas | |
| 5 | 57 | F | 1 | CRSsNP | N | Routine only | Y | Y | Y | Pseudomonas | |
| 6 | 63 | F | 2 | CRSwNP | Y | Routine & Pyro | Y | Y | Y | Pseudomonas | Serratia marcescens, Serratia,nematodiphila, Stenotrophomonas maltophilia, dickeya chrtsanthemi, enterobacterm staph aureus,citrobacter |
| 7 | 58 | F | 0 | CRSsNP | Y | Routine only | Y | Y | Y | Pseudomonas | |
| 8 | 73 | F | 1 | osteoma | N | Routine & Pyro | Y | Y | Y | Pseudomonas | Staphylococcus aureus, Staphylococcus lugdunensis |
| 9 | 68 | F | 2 | erting papillo | N | Routine only | Y | Y | Y | Pseudomonas | |
| 10 | 74 | M | 3 | erting papillo | N | Routine & Pyro | Y | Y | Y | Pseudomonas | Prevotella bivia |
| 11 | 53 | M | 2 | CRSwNP | N | Routine only | Y | Y | Y | ||
| 12 | 83 | M | 0 | CRSwNP | N | Routine only | Y | Y | Y | Alcaligenes faecalis,Pseudomonas aeruginosa | |
| 13 | 61 | M | 1 | CRSwNP | N | Routine only | Y | Y | Y | Staphylococcus aureus | |
| 14 | 83 | M | 1 | carcinoma | N | Routine only | Y | Y | Y | Staphylococcus aureus, Pseudomonas, aeruginosa | |
| 15 | 72 | F | 1 | CRSwNP | Y | Routine only | Y | Y | Y | ||
| 16 | 53 | F | 1 | CRSwNP | Y | Routine only | Y | Y | Y | Acromobacter xylosoxidans | |
| 17 | 67 | F | 3 | CRSsNP | Y | Routine only | Y | Y | Y | ||
| 18 | 73 | M | 4 | CRSwNP | Y | Routine & Pyro | Y | Y | Y | Pseudomonas aeruginosa | Staphylococcusylococcus aureus, Pseudomonas aeruginosa, Corynebacterium, propinquum |
| 19 | 72 | F | 2 | CRSsNP | N | Routine only | Y | Y | Y | ||
| 20 | 75 | F | 2 | CRSsNP | Y | Routine only | Y | Y | Y | ||
| 21 | 33 | F | 1 | CRSwNP | Y | Routine & Pyro | Y | Y | Y | Raoultella ornithinolytica, Corynebacterium tuberculostearicum,Klebsiella pneumoniae | |
| 22 | 57 | M | 1 | CRSsNP | N | Routine only | Y | Y | Y | ||
| 23 | 47 | M | 2 | CRSwNP | N | Routine only | Y | Y | Y | ||
| 24 | 76 | M | 0 | CRSsNP | N | Routine only | Y | Intermediate | N | ||
| 25 | 68 | M | 2 | erting papillo | N | Routine only | Y | Y | Y | Alcaligenes faecalis, Baceroides fragilis | |
| 26 | 26 | M | 1 | CRSwNP | N | Routine only | Y | Y | Y | ||
| 27 | 67 | M | 2 | CRSsNP | N | Pyroseq only | Y | Not tested | Y | Klebsiella | Haemophilus influenzae, Pneumoniae, Streptococcus agalactiae, klebsiella,Streptococcus pyogenes, Moraxella catarrhalis, Pseumdomanas aeruginosa,Staphylococcus aureus, pneumoniae, Streptococcus |
| 28 | 55 | F | 3 | CRSsNP | Y | Pyroseq only | Y | Not tested | Y | coagulase negative Staphylococcus | Haemophilius influenza, Pneumonia, Streptococcus agalactiae, klebsiella,Streptococcus pyogenes, Moraxella catarrhalis, Pseumdomonas aeruginosa,Staphylococcus aureus, streptococcus, Pneumonia, Streptococcus pneumoniae |
| 29 | 55 | M | 1 | CRSsNP | Y | Pyroseq only | Y | Not tested | Y |
Table 2: Combined patient data.
Discussion
To our knowledge, this is the only series describing a group of sinus disease patients with Serratia marcescens positive cultures of their purulence. Though there is no control group for comparison, it is notable that all 26/29 (89.6%) patients had previous functional endoscopic sinus surgery prior to the positive Serratia marcescens cultures. The implications of this are unknown, but given Serratia marcescens history as a nosocomial infection it does raise the concern that Serratia Marcescens presence in CRS could represent a post surgical disease. In the three patients who grew Serratia marcescens prior to ever having FESS, two of them also grew Pseudomonas Aeruginosa while only one had Serratia marcescens alone. Interestingly, these 3 patients did not have DNA pyrosequencing performed as we do not typically employ this technique before primary FESS in our patients. Whether Serratia marcescens is a post-surgical disease or can be present as the dominant bacteria in CRS before surgery requires future investigation.
Furthermore, the presence of Serratia marcescens may be dependent on the culture techniques performed as not all of our patients’ routine cultures grew Serratia marcescens. DNA-based molecular sequencing techniques have only recently been applied to the sinus microbiome; these techniques certainly offer expanded possibilities for investigation, but are not standard of care nor are they meant to replace standard culture techniques at this point. Our practice views them as complementary to routine culture techniques and employs the technology in recalcitrant cases of CRS.
Despite the lack of control group, it is important to highlight the high prevalence of co-morbid asthma in the CRS + Serratia marcescens patients as well as that greater than half (16/29 or 55%) of the patients had polymicrobial infections. This brings up the possibility that Serratia marcescens could play a role in the unified airway, similar to Pseudomonas aeruginosa [15].
The Serratia marcescens culture sensitivities for our patients at the University of Florida seem to support those found in the literature for other types of Serratia marcescens infections (12-14) with susceptibility to fluoroquinolones, trimethoprim-sulfamethoxazile, 3rd and 4th generation cephalosporins, piperacillintazobactam, ticarcillin-clavulanate, aztreonam, and carbapenems.
The high number of polymicrobial infections may be due to increased ability to find non-dominant organisms with DNA testing, and further research into this technique is required. When examining the treatment of Serratia marcescens with appropriate antimicrobials there does not seem to be a difference in symptomatic resolution in polymicrobials vs. unimicrobial infections following treatment. For both the unimicrobial and polymicrobial group there was a non-statistically different response rate (p=0.38) of 44.44% vs. 46.67%.
Similar to other published data Staphylococcus aureus and Pseudomonas aeruginosa were the most common polymicrobial isolates in our series [3]. Given that Staphylococcus Aureus has significant resistance to fluoroquinolones and high susceptibility to bactrim [16,17], Bactrim is likely a better choice in Serratia Marcecens polymicrobial infections with Staphylococcus aureus. This is opposed to Pseudomonas aeruginosa which is not bactrim sensitive but has susceptibility to fluoroquinolones such as ciprofloxacin and levofloxacin [18].
In patients with Serratia Marcecens who were appropriately treated and had follow up cultures 57% did not grow Serratia Marcencens on repeat culture with 43% did have persistent growth following treatment. This phenomenon is possibly explained by Serratia marcescens ability to create biofilms [19]. Although patient’s typically received 14- 21 days of therapy, Serratia marcescens ability to create biofilms may have protected it from being fully eradicated inspite of being susceptible to antimicrobial therapy.
This study certainly has limitations. First, it is retrospective and a small sample size. Secondly, there is no control group. Finally, there is heterogeneity in the culture techniques that were positive Serratia marcescens as well as the treatment regimens. More research into the utility of various culture techniques is needed. No valid conclusions could be drawn with regards to treatment given the variability of the treatments administered. Our goal of this study was to provide an initial characterization of sinus disease patients with Serratia Marscescens cultures.
Conclusion
This is the first literature to examine the characteristics of patients with Serratia marcescens in sinus disease patients to our knowledge. Serratia marcescens infection in the sinuses may be a post surgical disease, and it appears Levofloxacin, Ciprofloxacin, and TMP/SMX are all oral anti-microbials to which Serratia marcescens is consistently susceptible. The extent to which Serratia marcescens an instigator of recalcitrant disease or a bystander is unclear at this time and warrants further investigation.
References
- Rosenfeld RM, Piccirillo JF, Chandrasekhar SS, Brook I, Ashok Kumar K, et al. (2015) Clinical practice guideline (update): Adult sinusitis. Otolaryngol Head Neck Surg 152: 1-39.
- Larson D, Han J ( 2011) Microbiology of sinusitis: does allergy or endoscopic sinus surgery affect the microbiological flora. Current opinion in Otolaryngology & Head and Neck Surgery 19: 199-203.
- Brook I (2016) Microbiology of chronic rhinosinusitis. European Journal of Microbiology and Infectious Disease 35: 1059-1068.
- Hejazi A, Falkiner FR (1997) Serratia marcescens. J Med Microbiol 46: 903-912
- Mahlen SD (2011) Serratia infections: from military experiments to current practice. Clin Microbiol Rev 24: 755-791
- Anderson M, Stokken J, Sanford T, Aurora R, Sindwani R (2016) A systematic review of the sinonasal microbiome in chronic ehinosinusitis. Am J Rhinol Allergy 30: 161-166.
- Aurora R, Chatterjee D, Hentzleman J, Prasad G, Sindwani R, et al. (2013) Contrasting the microbiomes from healthy volunteers and patients with chronic rhinosinusitis. JAMA Otolaryngol Head Neck Surg 139: 1328-1338.
- Boase S, Foreman A, Cleland E, Tan L, Kreft RM, et al. (2013) The microbiome of chronic rhinosinusitis: culture, molecular diagnostics and biofilm detection. BMC Infectious Diseases 13: 210.
- Lee JT, Frank DN, Ramakrishnan V (2016) Microbiome of the paranasal sinuses: Update and literature review. Am J Rhinol Allergy 30: 3-16.
- Cope EK, Goldberg AN, Pletcher SD, Lynch SV (2017) Compositionally and functionally distinct sinus microbiota in chronic rhinosinusitis patients have immunological and clinically divergent consequences. Microbiome 5: 53.
- Dowd SE, Callaway TR, Wolcott RD, Sun Y, Mc Keehan T, et al. (2008) Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol 8: 125.
- Mahlen S, Morrow S, Abdalhamid B, Hanson ND (2003) Analyses of ampC gene expression in serratia marcescens reveal new regulatory properties. J Antimicrob Chemother 51: 791-802.
- Stock I, Grueger T, Wiedemann B (2003) Natural antibiotic susceptibility of strains of Serratia marcescens and the S. liquefaciens complex: S. liquefaciens sensu stricto, S. proteamaculans and S. grimesii. Int J Antimicrob Agents 22: 35-47.
- Michael J, Dowzicky MS, Park CH (2008) Update on antimicrobial susceptibility rates among gram negative and gram-positive organisms in the united states: Results from the tigecycline evaluation and surveillance trial (TEST) 2005 to 2007. Clinical Therapeutics 30.
- Illing E, Woodworth B (2014) Management of upper airway in cystic fibrosis. Curr Opin Pulm Med 20: 623-631.
- Gade N, Qazi M (2013) Fluoroquinolone therapy in Staphylococcus aureus infections: Where do we stand. Journal of Laboratory physicians 5: 109-112.
- Liu C, Bayer A, Cosgrove SE, Daum RS, Fridkin SK, et al. (2011) Management of patients with infections caused by methicillin-resistant staphylococcus aureus: Clinical practice guidelines by the infectious diseases society of america (IDSA). Clin Infect Dis 52: 18-55.
- Kanj S, Kanafani Z (2011) Current concepts in antimicrobial therapy against resistant gram-negative organisms: Extended-spectrum Ã-Lactamase?producing enterobacteriaceae, carbapenem-resistant enterobacteriaceae, and multidrug-resistant pseudomonas aeruginosa. Mayo Clin Proc 83: 250-259.
- Rice SA, Koh KS, Queck SY, Labbate M, Lam KW, et al. (2005) Biofilm formation and sloughing in serratia marcescens are controlled by quorum sensing and nutrient cues. J Bacteriol 187: 3477-4385.