Research Article - Biomedical Research (2017) Volume 28, Issue 9
The role of miR-139-5p in non-small cell lung cancer (NSCLC) with inactivation of HCCS1 gene
Zhifeng Lin1, Liwen Xiong2, Qiang Lin1*, Ying Chen1 and Xiangnan Xu11Department of Thoracic Surgery, Shanghai First People’s Hospital, Shanghai Jiaotong University, Shanghai, China
2Department of Pulmonary Medicine, Shanghai Chest Hospital, Shanghai Jiaotong University, 241 West Huaihai Road, Shanghai, China
- *Corresponding Author:
- Qiang Lin
Department of Thoracic Surgery
Shanghai First People’s Hospital
Shanghai Jiaotong University
China
Accepted date: February 07, 2017
Abstract
Objective: This research was to study the inactivation of HCCS1 gene in Chinese patients with Non- Small Cell Lung Cancer (NSCLC) and the mechanism of MicroRNA 139-5p in the inactivation process.
Methods: 51 clinical cases with non-small cell lung cancer were collected and were detected by immunohistochemistry, to study the expression of HCCS1 (hVPS53) protein and MicroRNA 139-5p in 51 cases of NSCLC carcinoma tissue and carcinoma side normal tissue, and the relationship between HCCS1 (hVPS53) expression levels and clinical pathological analysis the characteristics of NSCLC, the relationship between the and the expression of MicroRNA 139-5p and HCCS1 (hVPS53).
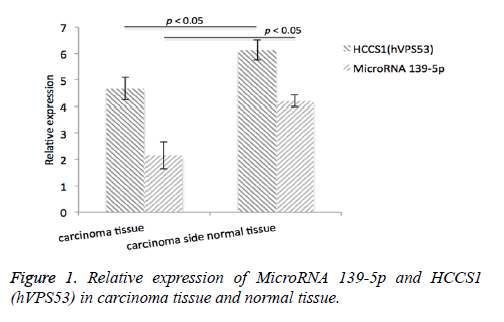
Results: Immunohistochemistry analysis of HCCSI (hVpS53) and MicroRNA39-5p levels in 51 carcinoma tissues. We found that HCCSI (hVpS53) protein expression level in carcinoma tissue was 4.68 ± 1.15 (relative expression), and 6.14 ± 1.24 (relative expression) in the carcinima side normal tissue. The expression level of MicroRNA39-5p in carcinoma tissue was 4.21 ± 0.38, and in the carcinima side normal tissue, the expression level was 2.15 ± 0.22. Analysis of HCCS1 (hVPS53) and MicroRNA 139-5p protein, their expression had significant difference, however none difference correlated with gender, age and pathological type, TNM staging and tumor grade (P>0.05), however there was a significant difference in the TNM stage and tumor type.
Conclusion: As a tumor suppressor gene, HCCS1 (hVPS53) gene has abnormal changes in many tumor tissues. This study shows that the MicroRNA 139-5p methylation acted on Chinese HCCS1 non-small cell lung cancer gene suppression caused by the loss of heterozygosity and abnormal methylation of gene silencing, may be the mechanism of HCCS1 gene inactivation.
Keywords
MicroRNA 139-5p, HCCS1 (hVPS53), NSCLC, Gene inactivation
Introduction
Lung cancer is the world's first cancer related death factors in non-small cell lung cancer (non-small cell lung cancer, NSCLC accounted for about 80%-85% of all lung cancer, smoking, environmental pollution etc. constituted the main reason for lung cancer. More than 40% of patients with NSCLC were treated with tumor recurrence in the early stage of treatment, and the survival rate of patients with advanced NSCLC was less than 5 and the prognosis was poor [1-3].
RNA (microRNA, miRNA) can regulate the expression of negatively in the post transcriptional level [4], participate in the occurrence and development of many kinds of tumors. MicroRNA plays an important role in the metastasis and invasion of tumor cells [5-7]. The specific role of microRNA-139-5p (miR-139-5p) in the development of lung cancer and the related mechanisms are not very clear. The short arm of human chromosome 17 has a high frequency of loss of heterozygosity in a variety of tumors, HIC-1 and other tumor suppressor genes have been cloned from this site [8], 17p13.3. In 2001, the 17p13.3 gene was cloned for the first time, which was named HCCS1. In this paper, we analysed the inactivation of HCCS1 gene in Chinese Non-Small Cell Lung Cancer (NSCLC) and the mechanism of MicroRNA 139-5p in the inactivation process of HCCS1.
Materials and Methods
51 cases of lung cancer tissue samples in this study were collected from April 2015 December~2015 surgical resection and postoperative pathological diagnosis were diagnosed as NSCLC patients, the clinical characteristics of patients are shown in Table 1 for each patient from the cancer tissues and the tissues of 2 copies, a rapidly placed liquid nitrogen freezing, another place in the formalin fixed. All patients were not treated with chemotherapy or radiotherapy before operation.
| Parameter | n | HCCS1 (hVPS53) | P | MicroRNA 139-5p expression | P |
|---|---|---|---|---|---|
| Gender | |||||
| Male | 24 | 4.58 ± 0.36 | 0.736 | 2.12 ± 0.45 | 0.637 |
| Female | 35 | 4.30 ± 0.41 | 1.94 ± 0.51 | ||
| Age/year | |||||
| ≤ 60 | 28 | 4.60 ± 0.39 | 0.933 | 2.15 ± 0.44 | 0.965 |
| >60 | 23 | 4.64 ± 0.45 | 1.88 ± 0.56 | ||
| Pathological type | |||||
| Squamous cell carcinoma | 31 | 4.61 ± 0.47 | 0.015 | 2.33 ± 0.47 | 0.031 |
| Adenocarcinoma | 12 | 3.28 ± 0.42 | 1.82 ± 0.41 | ||
| TNM stage | |||||
| I-II | 36 | 4.45 ± 0.46 | 0.024 | 2.12 ± 0.57 | 0.042 |
| III-IV | 15 | 2.37 ± 0.25 | 1.94 ± 0.42 | ||
| Tumor grade | |||||
| High-middle | 21 | 4.63 ± 0.46 | 0.785 | 2.10 ± 0.38 | 0.877 |
| Low | 30 | 3.97 ± 0.57 | 2.13 ± 0.40 | ||
Table 1: Relationship between the expression of HCCS1 (hVPS53), MicroRNA 139-5p protein and clinical pathological parameters.
Total RNA extraction and cDNA synthesis
The total RNA was extracted from cancer tissues and adjacent tissues according to Triblue specification [9]. 50 mg Triblue 1 ml were added to tissue, homogenized, kept in room temperature for 5 min, added chloroform 200 μl, 13400 Xg 4ºC centrifugal for 10 min, new 1.5 ml to learn from the upper water pipe, isopropanol. 3 times of volume (pre cooling) were added and then mixed, placed on ice 20 min. 4ºC, 13400 Xg centrifugal for 15 min. Discard the supernatant with 75% is 200 μL rinse for 1 times, fast dry centrifugal liquid, drying after adding sterile water DEPC 50 L dissolved RNA, according to the operation of subsequent experimental procedures or the remaining RNA save -80˚C. The total RNA was quantified by UV spectrophotometer.
Design of primers
According to the GenBank gene sequence, primers were designed with Primer 5 software [10]. HCCS1 (hVPS53) primer: 5'-CTGCACAGACTGAGTTAGGACA-3', primer: 5'- TCTCGTAGAACATTGCTGGGT-3', the length of the amplified products of 103 bp; housekeeping gene GAPDH upstream primer: 5’-GGTGAAGGTCGGAGTCAACGGA-3’, primer: 5’-GAGGGATCTCGCTCCTGGAAGA-3’, amplified length is 240 bp.
PCR
HCCS1 (hVPS53) gene was amplified by real-time fluorescence in IQ5 (Bio-Rad- Co., Ltd) fluorescence PCR instrument [11]. PCR process [11]. 10 μL was taken from the PCR reaction products, 2 μL 6X loading buffer was added and mixed into the sample of the 2% agarose gel, observed under UV lamp observation and took a picture.
Protein expression of HCCS1 (hVPS53) and MicroRNA 139-5p
Envision method was used; first, it was treated with poly-lysine slides, continuous paraffin sections of 4 μm, dried for 1 h. All the process was referenced from [12].
Relationship between HCCS1 (hVPS53), MicroRNA 139-5p and clinicopathological features
SPSS 13 software was used to analyse the relationship between HCCS1 (hVPS53) and MicroRNA 139-5p and clinicopathological features.
Statistics analysis
SPSS 13 statistical software was used for statistical analysis. After the normality test, paired t test was used to analyse the difference of HCCS1 (hvPS53) mRNA expression and the protein expression level between the cancer tissue and the adjacent lung tissue. Chi square test was used to analyse the relationship between the expression level of two different mRNA and clinical pathological parameters (α=0.05).
Results
Expression of HCCS1 (hVPS53) and MicroRNA 139-5p
Immunohistochemistry analysis of HCCSI (hVpS53) as well as MicroRNA39-5p levels in 51 carcinoma tissue tissues was conducted. We found that HCCSI (hVpS53) protein expression level in carcinoma tissue was 4.68 ± 1.15 (relative expression), and 6.14 ± 1.24 (relative expression) in the carcinoma side normal tissue. The expression level of MicroRNA39-5p in carcinoma tissue was 4.21 ± 0.38, and in the carcinoma side normal tissue, the expression level was 2.15 ± 0.22, Figure 1.
Relationship between the expression of HCCS1 (hVPS53), MicroRNA 139-5p protein and clinical pathological parameters
The protein levels of HCCS1 (hVPS53) as well as MicroRNA 139-5p have no significant difference with gender, age and pathological type, TNM staging and tumor grade (P>0.05), but there was a significant difference with TNM stage and tumor type. The expression of TNM in squamous cell carcinoma was higher than that in adenocarcinoma.
Discussion
In this study, we analysed the relationship between the inactivation of HCCS1 and MicroRNA 139-5p in non-small cell lung cancer cells. In this research, we utilized several experimental techniques to detect the changes of HCCS1 gene expression in primary lung cancer tissues. The results showed that the majority of tumor tissues did not express HCCS1 protein, and HCCS1 methylation in the promoter region of LOH was the main mechanism of HCCS1 gene inactivation. Interestingly, we found that the level of MicroRNA 139-5p has the same tendency with HCCS1 changes.
HCCS1 (hVPS53) protein was found in the search of tumor suppressor genes, which has high homology with the homologous protein of Arabidopsis, yeast, nematodes, fruit flies and mice [13]. It was very conservative in evolution and plays a very important role in the process of life. When the human hepatoma cells were induced, the tumor cells were obviously inhibited and the tumor cells were inhibited in nude mice. Observation in hepatocellular carcinoma was detected in HCCS1 (hVPS53) expression was significantly lower than that of the adjacent tissues [14], liver specimens of HCCS1 (hVPS53) gene mutation, single nucleotide deletion, the deletion of exon deletion and localization caused by reading frame change. Overexpression of hVPS53 (HCCS1) with HCCS1 (hVPS53) gene replication defective recombinant adenovirus Ad-HCCS1 mediated high expression [15]. Cell viability test showed that HCCS1 (hVPS53) protein could significantly inhibit the growth of human hepatoma cells and human colon cancer cells [16].
This is the first study using RT-PCR and immunohistochemistry were used to detect the tumor suppressor gene HCCS1 on chromosome 17p13.3 (hVPS53) and MicroRNA 139-5p protein level in NSCLC tissues and adjacent cancer tissues, HCCS1 (hVPS53) mRNA and MicroRNA 139-5p protein levels were lower than adjacent tissues, the difference was statistically significant (p<0.05).
The normal tissue had higher levels of HCCS1 (hVPS53), and cancer tissue had lower levels of HCCS1 (hVPS53), also think that HCCS1 (hVPS53) is a protective factor, this study further found that MicroRNA 139-5p in this process have the same phenomenon. It was found that the methylation level of MicroRNA 139-5p in cancer tissues was much higher than that in normal tissues. Therefore, inactivation of HCCS1 (hVPS53) gene may be related to the methylation of MicroRNA 139-5p. At the same time, the difference of mRNA and protein levels in cancer tissues and adjacent tissues was related to lymph node metastasis. The mRNA and protein levels of tumor with lymph node metastasis were lower than those in adjacent tissues (p<0.05). In this study, the number of cases of squamous cell carcinoma was much higher than that of adenocarcinoma and the relationship between the changes of HCCS1 (hVPS53) mRNA and protein level and the pathological type of tumor were not observed. The relationship between TNM and tumor grade and tumor grade was not observed. In addition, due to the limited number of samples, we need to analyse more samples to clarify the potential relationship among them.
Acknowledgements
This study was supported by Science Foundation of Shanghai Municipal Commission of Health and Family Planning (Grant No.20124350).
References
- Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer 2003; 3: 55-63.
- Tran YK, Bogler O, Gorse KM, Wieland I, Green MR. A novel member of the NF2/ERM/4.1 superfamily with growth suppressing properties in lung cancer. Cancer Res 1999; 59: 35-43.
- Choudhury A, Palma M, Mellstedt H. The future of cancer vaccines for non-small-cell lung cancer: ongoing trials. Clin Lung Cancer 2008; 9: 37-44.
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001; 411: 494-498.
- Ribeiro-Filho LA, Franks J, Sasaki M, Shiina H, Li LC. CpG hypermethylation of promoter region and inactivation of E-cadherin gene in human bladder cancer. Mol Carcinog 2002; 34: 187-198.
- Zhang SH, Cong WM, Xian ZH, Wu MC. Clinico pathological significance of loss of heterozygosity and microsatellite instability in hepatocellular carcinoma in China. World J Gastroenterol 2005; 11: 3034-3039.
- Shinji K, Daisuk Y, Takeshi F. Promoter methylation of DAL-l/4.IB predicts poor prognosis in non-small cell lung cancer. Clin Cancer Res 2005; 2954-2961.
- Wittmann T, Bokoch GM, Waterman-Storer CM. Regulation of microtubule destabilizing activity of Op18/stathmin downstream of Rac1. J Biol Chem 2004; 279: 6196-6203.
- Guerrera F, Tabbn F, Bessone L, Maletta F. The influence of tissue ischemia time on rna integrity and patient-derived xenografts (PDX) engraftment rate in a non-small cell lung cancer (NSCLC) Biobank. PLoS One 2016; 11: e0145100.
- Xiao-Yong S, Zhi-Feng L, Fan-Zhen L, Zhen R, Jian Z. Expression and clinical significance of HCCS1 in non-small cell lung cancer. Contemp Oncol (Pozn) 2012; 16: 328-331.
- Xu W, Hang M, Yuan CY. MicroRNA-139-5p inhibits cell proliferation and invasion by targeting insulin-like growth factor 1 receptor in human non-small cell lung cancer. Int J Clin Exp Pathol 2015; 8: 3864-3870.
- Zhifeng L, Hailong H, Chaoqiang Q. Expression and clinical significance of HCCS1(hVPS53) in NSCLC patients. China Oncol 2011; 11: 841-845.
- Messenguy F, Dubois E. The yeast ARGRII regulatory protein has homology with various RNases and DNA binding proteins. Mol Gene Genom 1988; 211: 102-105.
- Xie YEC, Sheng J. Effect of Cdc42 gene inhibited on proliferation, migration and invasion in human hepatocellular carcinoma cells. Zhonghua Wai Ke Za Zhi Chin J Surg 2015; 53: 957-962.
- Zheng HF, Bai ZY, Lin JY. Characterization and functional analysis of a chitin synthase gene (HCCS1) identified from the freshwater pearlmussel Hyriopsis cumingii. Gene Mol Res Gmr 2015; 14: 19264-19274.
- Murad LD, Soares NC, Brand C. Effects of caffeic and 5-caffeoylquinic acids on cell viability and cellular uptake in human colon adenocarcinoma cells. Nutr Cancer 2015; 67: 532-542.