Research Article - Biomedical Research (2017) Volume 28, Issue 11
The regulation of chronic inflammatory persistent pain by Dnmt3a in the spinal cord in rats
Shao Cuijie1*#, Gao Yong1#, Jin Dan1, Luan Shunlian2 and Wang Deqiang1,2*
1Department of Pain, the Affiliated Hospital of Binzhou Medical University, Binzhou, PR China
2Department of Oncology, the Affiliated Hospital of Binzhou Medical University, Binzhou, PR China
#These authors contributed equally to this work
- *Corresponding Authors:
- Shao Cuijie
Department of Pain
The Affiliated Hospital of Binzhou Medical University, PR China
Accepted date: March 28, 2017
Abstract
Purpose: Persistent inflammatory or neuropathic pain is common in clinical practice. DNA methylation is reported to be involved in chronic pain, but its function has not been fully investigated. Thus, our study aimed to investigate the role of Dnmt3a in the superficial dorsal horn in regulating the maintenance phase of persistent pain at cellular and behavioral levels.
Methods: CFA-induced chronic pain adult male SD rat model was established by injecting CFA into the left talocrural joint. Thermal Withdrawal Latency (TWL) and Mechanical Withdrawal Threshold (PWT) were used for determining the pain degree. Real-time RT-PCR and western blot were used to detect the DNA and protein expression. The specimen of lumbar 4-6 spinal cords was collected.
Results: DNA methylation and DNA methyltransferase Dnmt3a activity in the superficial dorsal horn was associated with chronic inflammatory pain in CFA rat model and Dnmt3a expression was upregulated. Intrathecal Dnmt3a-Pr aggravated hyperalgesia, while intrathecal Dnmt3a-siRNA reversed the chronic inflammatory persistent pain. Our in vivo data indicated that Dnmt3a-siRNA could alleviate inflammatory pain.
Conclusion: Dnmt3a could participate in the mediation of chronic persistent pain at cellular and behavioral levels, which may be identified as a new therapeutic agent for treating such disease.
Keywords
Inflammatory persistent pain, Epigenetic mechanisms, Dnmt3a, Rat superficial dorsal horn, Methylation.
Highlights
DNA methylation participate in regulate chronic inflammatory pain. Dnmt3a regulate DNA methylation in rat spinal cord. Dnmt3a-siRNA alleviate inflammatory pain.
Abbreviations
DNMTs: DNA Methyltransferases; CFA: Complete Freund’s Adjuvant; TWL: Thermal Withdrawal Latency; PWT: Paw Withdrawal Threshold.
Introduction
Persistent inflammatory or neuropathic pain is a very common disease in clinical practice. The induction and maintenance of persistent inflammatory or neuropathic pain involve a number of variations in gene expression in the spinal cord [1,2]. More stable epigenetic modifications [3], such as DNA methylation and histone acetylation, can more persistently affect gene expression in the spinal cord to alter the function. DNA methylation, an epigenetic mechanism catalysed by DNA Methyltransferases (DNMTs), is involved in many cellular processes [4], and has recently been implicated in memory formation [5]. Since pain pathogenesis and memory formation are both associated with synaptic plasticity [6], it is likely that DNA methylation as well as consequent DNMT activity also participate in the development of pain [7].
Despite the abundant neuronal expression of Dnmts [8], little is known regarding the function of DNA methylation in the spinal cord. Recently, we recognized a crucial role of Dnmt3a activity in the pattern of gene expression in the spinal cord subsequent to injection of Complete Freund’s Adjuvant (CFA) into the rat talocrural joint, followed by a rapid increase in Dnmt3a expression. Notably, the potential role for neuronic Dnmt3a needs further lucubration. In the present study, we examined the spinal cord to determine whether these concepts regarding DNA methylation could extend to CFA models. Toward this goal, we focused on the specific Dnmt3a that was regulated in chronic pain, and the function of DNA methylation in the spinal cord that affected persistent pain sensation.
Materials and Methods
Talocrural joint inflammation animal model
Adult male Sprague-Dawley rats (6-8 weeks) used in this experiment were provided by the experimental animal center of Binzhou Medical University. Animals were housed to acclimate for 1 week prior to experimentation on a 12 h lightdark cycle with ad libitum access to food and water. All experimental protocols were approved by the Animal Care and Use Committee of Binzhou Medical University (Binzhou, Shandong, China) and according to the Declaration of National Institutes of Health Guide for Care and Use of Laboratory Animals (Publication No. 80-23, revised 1996).
Inflammation was induced by injection of 10 μl Complete Freund’s adjuvant (CFA, Sigma) in the left talocrural joint. Animals were anesthetized with 2% isoflurane in an enclosed chamber. The needle was anterolaterally inserted into the ankle joint, with the ankle fixed in plantar flexion to expose the joint. Sham treatment consisted of anesthesia and injection of 10 μl normal saline in the left ankle joint. At 4 h, 3 d and 14 d after CFA injection, six rats from each group were randomly allotted for global DNA methylation with ELISA, Dnmt3a mRNA expression with RT-PCR, and protein detection with western blot analysis. The rats were sacrificed under deep anesthesia, with their lumbar 4-6 spinal cords immediately isolated.
Dnmt3a overexpression and silencing
For Dnmt3a overexpression, we used Dnmt3a-active protein fragments (Ambion), which directly increased the content of Dnmt3a. In this system, due to intrathecal injection to spinal cord Dnmt3a occurred selectively in spinal cord. To inhibit the Dnmt3a expression, we introduced a vector containing the Dnmt3a-siRNA sequence as 5’- CAGGAGAUGAUGUCCAACCC-3’ (Ambion). The Dnmt3atargeting siRNA sequence recognized the 5’-UTR sequence unique to the Dnmt3a sequence and inhibited Dnmt3a gene transcription. The levels of Dnmt3a in the spinal cord of CFA rats was manipulated by intrathecal injection of Dnmt3a-active protein fragments or delivered Dnmt3a-siRNA.
Neuropathic pain measurements of thermal hyperalgesia and mechanical allodynia
Ipsilateral thermal hyperalgesia was measured by the IITC Plantar Analgesia Meter (IITC Life Science Inc., Victory Blvd Woodland Hills, CA, USA) for thermal Withdrawal Latency (TWL) according to the method described [8]. In brief, rats were placed in transparent acrylic enclosures with a glass plate (22 × 12 × 12 cm), and allowed to acclimatize to their environment for 30 min before testing in a temperaturecontrolled and noise-free room. The high-intensity, movable radiant heat source was placed underneath the glass and focused onto the plantar surface of each hind paw. The nociceptive endpoint in the radiant heat test was characterized by lifting of the hind paw. The time from the onset of radiant heat to the endpoint was considered as TWL. The radiant heat intensity was adjusted at the beginning of the experiment to obtain basal TWL of 12-15 s, and kept constant thereafter. An automatic 20 s cut-off was used to prevent tissue damage. Each animal was tested twice on each hind paw at an interval of 5 min. TWLs of each rat were measured 3 days before CFA injection, and at time points of 0, 4 h, 1 d, 3 d, 5 d, 7 d and 14 d after CFA surgery. All behavioral tests were conducted by observers blinded to the animal treatments.
Ipsilateral mechanical allodynia was assessed with electronic von Frey filaments (IITC Life Science Inc., Victory Blvd Woodland Hills, CA, USA). Animals were placed in individual plastic boxes (30 × 20 × 15 cm) on a metal mesh floor and allowed to acclimate for 30 min. The filaments were presented, perpendicular to the plantar surface with sufficient force to cause slight bending against the paw and held for 1-2 s. Brisk withdrawal or paw flinching was considered as positive response. The Paw Withdrawal Threshold (PWT) was determined with the digital screen display. Each animal was tested in triplicate on each hind paw at an interval of 30 s. PWTs of each rat were measured 3 days before CFA, and at 0 d, 4 h, 1 d, 3 d, 5 d, 7 d and 14 d after CFA surgery. All behavioral tests were performed by two independent observers.
Global DNA methylation analysis
We used Epigentek’s Methylamp Global DNA Methylation Quantification Ultra Kit in accordance with the manufacturer’s instructions to determine the level of global DNA methylation. Raw values were colorimetrically quantified with ELISA and total methylation levels estimated by generation of a standard curve from Epigentek’s methylated DNA standard. Values are represented as methylation percentage relative to vehicle control. The following steps: prepare DNA, adding 2 μl (100-200 ng) of sample DNA and 98 μl solution (kit support); shake the plate frame to allow the solution to cover the entire surface of each well’s bottom. Incubate the wells at 37°C (with no humidity) for 40 min, followed by incubation at 60°C (with no humidity) for 35-40 min to evaporate the solution and dry the wells. Make sure that the well was completely dry by slightly tilting the well and aspirating against the edge with a P-10 or P-20 pipette. Add 150 μl solution to each dried well. Incubate at 37°C for 30 min. Aspirate and wash each well with 150 μl of the diluted solution each time for 3 times. Add 100 μl solution to each well and incubate at room temperature for 1-5 min away from light. Add 50 μl solution to each well to stop enzyme reaction when the color in the standard wells containing the higher concentrations of standard control turned medium blue. The color will change to yellow and the absorbance should be read on a microplate reader at 450 nm within 2-15 min.
RNA isolation and real-time RT-PCR
Tissues for RNA isolation were obtained and rapidly placed in Trizol (Invitrogen, Carlsbad, CA, USA) and processed according to the manufacturer's instructions. Total RNA was purified with RNAesy Micro columns (Qiagen, Valencia, CA, USA) and spectroscopy confirmed that the RNA 260/280 ratio was between 1.8 and 2.0. For the generation of cDNA, total RNA was reverse-transcribed using the High-Capacity cDNA reverse transcription kit (Taraka, Dalian, China). Quantitative real-time PCR was performed using SYBR Green I (Taraka, Dalian, China) (Applied Biosystems Step-one TM Real-Time PCR System), and approximately 500 ng of cDNA per reaction. Each reaction was conducted in triplicate and quantified using the ΔΔCt method [8]. For the PCR amplification, DNMT3A-specific primers was performed with denaturation at 95°C for 10 min, followed by 35 denaturation cycles at 95°C for 20 s, primer annealing at 58°C for 20 s, and a primer extension phase at 72°C for 20 s. A final extension step at 72°C for 5 min was performed before the reaction mixture was stored at 4°C. The primer sequences were as follows: DNMT3A (sense) 5'- TCCTGCTGTGTGGTTAGACG-3', (antisense) 5'- TATTTCCGCCTCTGTGGTTT-3'; and GAPDH (sense) 5'- CTCCTCCTGTTCGACAGTCAG-C-3', (antisense) 5'- CCCAATACGACCAAATCCGTT-3'. Expression data were normalized to the geometric mean of GAPDH to control the variability in expression levels. In all cases, the validity of amplification was confirmed by the presence of a single peak in the melting temperature analysis and linear amplification with increasing number of PCR cycles.
Western blot
Total protein extracts from lumbar 4-6 spinal cord induced with Dnmt3a-active protein fragment or Dnmt3a-siRNA or Sham or CFA were prepared using the Protein Kit (Qiagen, Valencia, CA, USA) according to manufacturer’s instructions. Fresh spinal cord tissues and compared normal tissues were ground to powder in liquid nitrogen and lysed with SDS-PAGE sample buffer. For detection of DNMT3A expression in protein level, equal protein samples (30 μg) extracted from the tissues were loaded per lane, separated on 8% SDS-polyacrylamide gels and transferred to PVDF membranes (Immobilon P, Millipore, Bedford, MA). The membranes were blocked with 5% fat-free milk in Tris-buffered saline containing 0.1% Tween-20 (TBST) for 0.5 h at room temperature. The primary antibodies were Dnmt3a (1:500, Epigentek, UK) and GAPDH (1:5000, Sigma). The membranes were blocked and probed with antibodies to Dnmt3a (1:1000, Epigentek, UK), followed by incubation with horseradish peroxidase-conjugated secondary antibodies (1:5000) at room temperature on a shaker for 1 h. The immune complexes were visualized by chemiluminescence using an ECL kit (Pierce, USA). Positive bands were analysed with Quantity One software (Bio-Rad). Dnmt3a expression levels were normalized to GAPDH levels.
Statistical analysis
All the statistical analyses were performed using SPSS 17.0 software (SPSS Inc. Chicago, IL, the USA). Two-tailed unpaired Student’s t tests were used throughout this study to compare the continuous data between two distinct groups. One-way ANOVA was used for pharmacological verification of changes in global methylation. For mRNA and protein detection, Student’s t test was performed. All error bars represented S.E.M. P value less than 0.05 was considered to be statistically significant.
Results
Dnmt3a expression was upregulated in chronic pain rat model
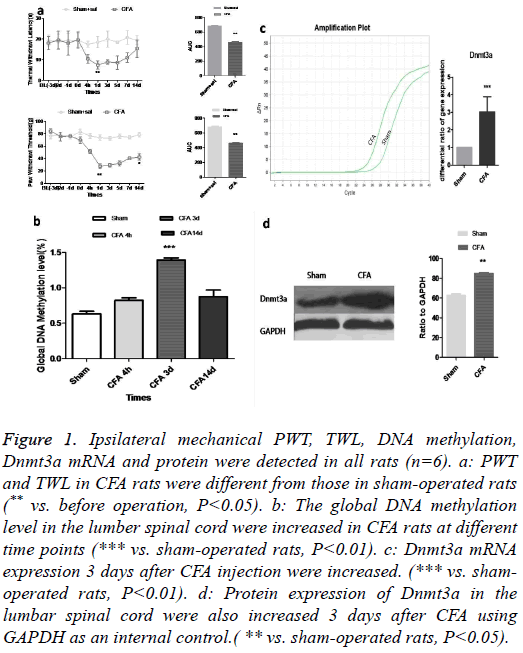
The CFA rat model was successfully established which was validated by measuring TWL and PWT (Figure 1a). To investigate the possible causality of DNA hypermethylation in CFA-induced persistent pain, we compared the levels of total DNA methylation in sham and CFA adult rats. We found that the levels of DNA methylation were increased in spinal cords (Figure 1b). Since DNA methylation is an epigenetic mechanism catalysed by DNMTs [9], we determined the levels of Dnmts transcripts using fluorogenic real-time RT-PCR, and the Dnmt3a transcripts were increased in the spinal cords of CFA adult rats (Figure 1c) and the Dnmt3a protein was correspondingly upregulated (Figure 1d).
Figure 1: Ipsilateral mechanical PWT, TWL, DNA methylation, Dnmt3a mRNA and protein were detected in all rats (n=6). a: PWT and TWL in CFA rats were different from those in sham-operated rats (** vs. before operation, P<0.05). b: The global DNA methylation level in the lumber spinal cord were increased in CFA rats at different time points (*** vs. sham-operated rats, P<0.01). c: Dnmt3a mRNA expression 3 days after CFA injection were increased. (*** vs. shamoperated rats, P<0.01). d: Protein expression of Dnmt3a in the lumbar spinal cord were also increased 3 days after CFA using GAPDH as an internal control.( ** vs. sham-operated rats, P<0.05).
The Dnmt3a genomic locus on 2p33, which is concentrated in heterochromatin [10], is considered to be transcriptionally silent and functions primarily as a transcriptional repressor [11]. We found that Dnmt3a served as a classical immediateearly gene [12], it was robustly and persistently activated by CFA-induced pain. Notably, pain-activated Dnmt3a was increased in the spinal cord in CFA rats at 4 hours and lasted till day 14 or longer.
Down-regulation of Dnmt3a reversed CFA-induced pain and spinal neuronal sensitization
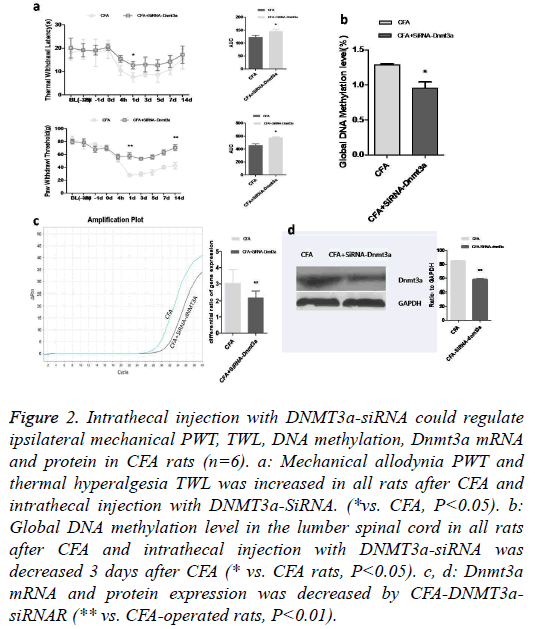
A pain-dependent increase in DNA methylation has been reported to be activated mainly in heterochromatin DNA sequences [13]. Given the association of Dnmt3a with heterochromatin and its robust regulation by neuronal activity [14], we investigated the increase of Dnmt3a in CFA rats as a potential mechanism. We speculated that decreasing the levels of Dnmt3a in the spinal cord in CFA rats would restore their pain threshold. To ascertain the association of increased Dnmt3a levels in the spinal cord in CFA rats with exacerbated pain intensities, we selectively manipulated the levels of Dnmt3a in the spinal cord of CFA rats by intrathecal injection of delivered Dnmt3a-siRNA. 3 days after CFA and Dnmt3asiRNA intrathecal injection, the increment percentage of analgesic threshold was observed (Figure 2a). We found that suppressed expression of Dnmt3a in the spinal cord of CFA rats induced a significant decrease in the global DNA methylation levels (Figure 2b). The primary afferent activity of the spinal cord is particularly associated with algesia, indicating that DNMTs play an important role in this pain process. We found that Dnmt3a mRNA and protein in CFA rats injected with Dnmt3a-SiRNA exhibited lower expression than CFA rats (Figures 2c and 2d), suggesting Dnmt3a-SiRNA suppressed the expression of Dnmt3a gene and alleviated the pain intensity. Together, these findings indicate that pain is associated with an increase in the expression of Dnmt3a and restoring the levels of Dnmt3a in the CFA rat’s spinal cords leads to recovery of pain threshold.
Figure 2: Intrathecal injection with DNMT3a-siRNA could regulate ipsilateral mechanical PWT, TWL, DNA methylation, Dnmt3a mRNA and protein in CFA rats (n=6). a: Mechanical allodynia PWT and thermal hyperalgesia TWL was increased in all rats after CFA and intrathecal injection with DNMT3a-SiRNA. (*vs. CFA, P<0.05). b: Global DNA methylation level in the lumber spinal cord in all rats after CFA and intrathecal injection with DNMT3a-siRNA was decreased 3 days after CFA (* vs. CFA rats, P<0.05). c, d: Dnmt3a mRNA and protein expression was decreased by CFA-DNMT3asiRNAR (** vs. CFA-operated rats, P<0.01).
Upregulation of Dnmt3a aggravates CFA-induced pain behavior and spinal neuronal sensitization
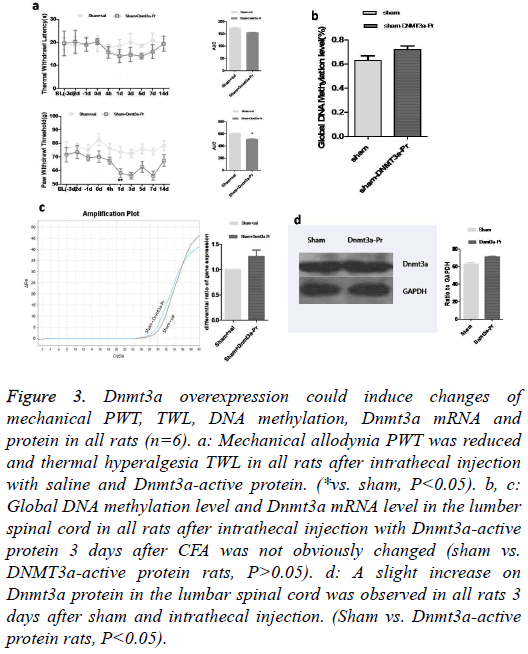
To further address the role of Dnmt3a in pain intensity, we investigated whether a increment in Dnmt3a levels in the spinal cords of normal rats, which would generate an ache-like phenotype with pain hypersensitivity. Dnmt3a-active protein was intrathecally injected into the spinal cords in the rats. Behavioral analysis using Plantar Analgesia Instrument and Mechanical Analgesia Instrument revealed that no changes were observed in TWL, but PWT was reduced (Figure 3a). Neither the global DNA-methylation (Figure 3b) nor the Dnmt3a-mRNA (Figure 3c) was affected. The Dnmt3a protein increased slightly (Figure 3d).
Figure 3: Dnmt3a overexpression could induce changes of mechanical PWT, TWL, DNA methylation, Dnmt3a mRNA and protein in all rats (n=6). a: Mechanical allodynia PWT was reduced and thermal hyperalgesia TWL in all rats after intrathecal injection with saline and Dnmt3a-active protein. (*vs. sham, P<0.05). b, c: Global DNA methylation level and Dnmt3a mRNA level in the lumber spinal cord in all rats after intrathecal injection with Dnmt3a-active protein 3 days after CFA was not obviously changed (sham vs. DNMT3a-active protein rats, P>0.05). d: A slight increase on Dnmt3a protein in the lumbar spinal cord was observed in all rats 3 days after sham and intrathecal injection. (Sham vs. Dnmt3a-active protein rats, P<0.05).
Thus, increased levels of Dnmt3a correlate with increased expression of DNA methylation, in consistence with the basis of the conventional notion that DNA methylation is associated with transcription repression [15]. However, the mechanisms of transcription regulation by DNA methylation appear to be more complex than previously conceived. DNA methylation can also be associated with transcription activation [16], consistent with our finding that Dnmt3a is required for specific, activity-driven genomic responses.
Our findings indicated that the levels of Dnmt3a in the spinal cord determined the ability to keep constant pain state, and that a pain induced increase on Dnmt3a expression is causally linked to pain amplification. These results suggested a gaining function of DNA methylation in pain maintenance and Dnmt3a is identified as a potential target for alleviating algogenia in CFA rats.
Discussion
Chronic inflammatory pain is characterized by hyperalgesia, allodynia and spontaneous pain [17]. The pathogenesis of inflammatory persistent pain is complicated, with many factors unknown. Recent evidence suggests that expression levels of pain-associated genes in sensory neurons are involved in the generation and maintenance of inflammatory pain [18]. Epigenetic mechanisms can regulate the transcription and expression of pro- or anti-nociceptive genes [19]. DNA methylation, one of the important means of epigenetic regulation, has recently been implicated in chronic persistent pain. In animal models of pain [20], the DNA methylation is intensified in many tissues, including the spinal cord [21]. This test is to investigate a possible causative link between DNA hypermethylation and chronic pain. Therefore, we hypothesized that DNA methylation and Dnmt3a activity may be involved in the pathogenesis and maintenance of inflammatory pain. In this study, we examined global DNA methylation and Dnmt3a expression in the spinal cord in rats following CFA surgery. We further examined the effects of intrathecal administration of the Dnmt3a-siRNA or Dnmt3aactive protein fragment to block or activate the expression of Dnmt3a, followed by detection of the above mentioned indexes. The differences between the normal and CFA rats were compared and analysed. All results elucidating that the role of DNA methylation in late chronic inflammatory persistent pain may facilitate development of new therapies. Target therapies are urgently needed because conventional painkillers such as opioids and Non-Steroidal Anti- Inflammatory Drugs (NSAIDS) have poor efficacy in this pain state [22]. It was long considered that mature neurons had no potential to synthesize DNA and neurogenesis was nonexistent in the adult mammalian Central Nervous System (CNS). Recent studies have shown that DNA synthesis and neurogenesis occur in the adult mammalian CNS under pathological conditions such as injury. Cell proliferation and DNA synthesis has been recognized in the glial cells in the adult mammalian CNS after injury [23]. Thus, DNA synthesis may occur in the neurons and glial cells in the spinal dorsal horn after nerve damage in CFA rats [24,25]. In our current study, CFA-induced mechanical allodynia and thermal hyperalgesia were markedly attenuated by intrathecal administration of Dnmt3a-siRNA from day 0 to day 2 in CFA rats. Moreover, intrathecal Dnmt3a-siRNA treatment may alleviate inflammatory pain by inhibiting the expression of Dnmt3a, with the expression of DNA methylation-dependent algogenic genes downregulated in the lumbar spinal cord in CFA rats, which might be attributed to reduced global DNA methylation and MeCP2 expression as the result of Dnmt3asiRNA intrathecal administration. The changes of behavior tests were obvious on Day 1, while the accumulation of expression and global DNA methylation were peaked on Day 3. Our subsequent research would be focused on the investigation of the exact mechanisms underlying the downregulation of MeCP2 expression via Dnmt3a-siRNA.
Patterns of DNA methylation are established and maintained by DNMTs [26]. While Dnmt1 is the maintenance methyltransferase which ensures that methylation patterns are faithfully copied throughout each cell division, with Dnmt3a and Dnmt3b regulating de novo DNA methylation in response to environmental factors [27,28]. DNMT expression is upregulated coupled with an upregulated MeCP2 [29]. One hypothesis is that the downregulated Dnmt3a leads to reduction of DNA methylation, so the synthesis of DNA methylation binding protein MECP2 is inhibited. Less MeCP2 may be needed to bind methyl-CpG, followed by decreased methyl- CpG in the nuclei [30]. Some pain-related genes, such as BDNF, which could be combined with MeCP2 protein, cannot be de facto bound to the right site, with the methylation of BDNF promoter reduced and the pain symptoms relieved. Despite the consistency of mechanical pain threshold, PWT or also called thermal pain threshold was reduced subsequent to intrathecal injection of Dnmt3a-active protein fragments in CFA rats, which might be attributable to the inconsistency of thermal and mechanical pain signal pathways.
Thus, DNA methylation may play a significant role in chronic inflammatory persist pain. Dnmt3a has been approved by the Ley and Drug Administration for chemotherapy against myelodysplastic syndrome [31] and some other malignant diseases such as acute myeloid leukemia [32]. It may also be a potential drug in the treatment of inflammatory persistent pain. However, this study had some limitations and was only focused on changes within 7 days after CFA. Further studies are required as to examine the long-term inflammatory persistent pain and DNA methylation as well as other models such as CCI.
In summary, our results suggest that DNA methylation and Dnmt3a may play an important role in inflammatory persist pain. Inhibition of DNMTs may be a new therapeutic approach for inflammatory persist pain and may provide some hints for further studies of the involvement of DNA methylation in pain areas.
Conflict of interest
None
Acknowledgements
This work was supported by Youth Fund of National Natural Science Foundation of China (Grant No. 81100813 to Shao Cuijie), Natural Science Fund for Colleges and Universities of Jiangsu Educational Department (Grant No. 11KJB320018 to Shao Cuijie), China Postdoctoral Science Foundation (Grant No. 2012M510189 to Shao Cuijie), Project of Shandong Province Higher Educational Science and Technology Program (Grant No. J14LL02), Youth Fund of the Affiliated Hospital of Binzhou Medical University (Grant No. 2013QNKYJJ09), and the Doctoral Startup Fund Program Funded by the Affiliated Hospital of Binzhou Medical College.
References
- Crown ED. Calcium/calmodulin dependent kinase II contributes to persistent central neuropathic pain following spinal cord injury. Pain 2012; 153: 710-721.
- Lozano-Ondoua AN, Symons-Liguori AM, Vanderah TW. Cancer-induced bone pain: Mechanisms and models. NeurosciLett 2013; 557: 52-59.
- Doehring A, Geisslinger G, Lotsch J. Epigenetics in pain and analgesia: an imminent research field. Eur J Pain 2011; 15: 11-16.
- Buchheit T, Van de Ven T, Shaw A. Epigenetics and the transition from acute to chronic pain. Pain Med 2012; 13: 1474-1490.
- Rahn EJ. Cellular, molecular, and epigenetic mechanisms in non-associative conditioning: implications for pain and memory. Neurobiol Learn Mem 2013; 105: 133-150.
- Jarome TJ, Lubin FD. Epigenetic mechanisms of memory formation and reconsolidation. Neurobiol Learn Mem 2014; 115: 116-127.
- Qiu S, Li XY, Zhuo M. Post-translational modification of NMDA receptor GluN2B subunit and its roles in chronic pain and memory. Semin Cell DevBiol 2011; 22: 521-529.
- LaPlant Q, Vialou V, Covington HE, Dumitriu D, Feng J. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci 2010; 13: 1137-1143.
- Turek-Plewa J, Jagodziaski PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell MolBiolLett 2005; 10: 631-647.
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science 2001; 293: 1089-1093.
- Wu H, Coskun V, Tao J, Xie W, Ge W. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 2010; 329: 444-448.
- Gray KS. The de novo methyltransferases DNMT3a and DNMT3b target the murine gammaherpesvirus immediate-early gene 50 promoter during establishment of latency. J Virol 2010; 84: 4946-4959.
- Doehring A, Oertel BG, Sittl R, Lotsch J. Chronic opioid use is associated with increased DNA methylation correlating with increased clinical pain. Pain 2013; 154: 15-23.
- Chestnut BA, Chang Q, Price A, Lesuisse C, Wong M. Epigenetic regulation of motor neuron cell death through DNA methylation. J Neurosci 2011; 31: 16619-16636.
- Spruijt CG, Vermeulen M. DNA methylation: old dog, new tricks? Nat StructMolBiol 2014; 21: 949-954.
- Gos M. Epigenetic mechanisms of gene expression regulation in neurological diseases. ActaNeurobiolExp (Wars) 2013; 73: 19-37.
- Carlton SM. Peripheral and central sensitization in remote spinal cord regions contribute to central neuropathic pain after spinal cord injury. Pain 2009; 147: 265-276.
- Honore P. Murine models of inflammatory, neuropathic and cancer pain each generates a unique set of neurochemical changes in the spinal cord and sensory neurons. Neuroscience 2000; 98: 585-598.
- Tochiki KK, Cunningham J, Hunt SP, Geranton SM. The expression of spinal methyl-CpG-binding protein 2, DNA methyltransferases and histone deacetylases is modulated in persistent pain states. Mol Pain 2012; 8: 14.
- Hwang CK, Song KY, Kim CS, Choi HS, Guo XH. Epigenetic programming of mu-opioid receptor gene in mouse brain is regulated by MeCP2 and Brg1 chromatin remodelling factor. J Cell Mol Med 2009; 13: 3591-3615.
- Deshpande A, Furlan A, Mailis-Gagnon A, Atlas S, Turk D. Opioids for chronic low-back pain. Cochrane Database Syst Rev 2007; 004959.
- Grinspan JB, Reeves MF, Coulaloglou MJ, Nathanson D, Pleasure D. Re-entry into the cell cycle is required for bFGF-induced oligodendroglial dedifferentiation and survival. J Neurosci Res 1996; 46: 456-464.
- Luzzati F, De Marchis S, Fasolo A, Peretto P. Neurogenesis in the caudate nucleus of the adult rabbit. J Neurosci 2006; 26: 609-621.
- Chen J, Magavi SS, Macklis JD. Neurogenesis of corticospinal motor neurons extending spinal projections in adult mice. ProcNatlAcadSci USA 2004; 101: 16357-16362.
- Talos F, Abraham A, Vaseva AV, Holembowski L, Tsirka SE. p73 is an essential regulator of neural stem cell maintenance in embryonal and adult CNS neurogenesis. Cell Death Differ 2010; 17: 1816-1829.
- Neary JT. Opposing effects of P2X (7) and P2Y purine/pyrimidine-preferring receptors on proliferation of astrocytes induced by fibroblast growth factor-2: implications for CNS development, injury, and repair. J Neurosci Res 2008; 86: 3096-3105.
- Bailey ZS. Blast induced neurotrauma causes overpressure dependent changes to the DNA methylation equilibrium. NeurosciLett 2015; 604: 119-123.
- Subramaniam D, Thombre R, Dhar A, Anant S. DNA methyltransferases: a novel target for prevention and therapy. Front Oncol 2014; 4: 80.
- Kinney SR, Pradhan S. Regulation of expression and activity of DNA (cytosine-5) methyltransferases in mammalian cells. ProgMolBiolTranslSci 2011; 101: 311-333.
- Anier K, Malinovskaja K, Aonurm-Helm A, Zharkovsky A, Kalda A. DNA methylation regulates cocaine-induced behavioral sensitization in mice. Neuropsychopharmacology 2010; 35: 2450-2461.
- Kyzar EJ, Pandey SC. Molecular mechanisms of synaptic remodeling in alcoholism. NeurosciLett 2015; 601: 11-19.
- Jhanwar SC. Genetic and epigenetic pathways in myelodysplastic syndromes: A brief overview. AdvBiolRegul 2015; 58: 28-37.