Research Article - Biomedical Research (2018) Volume 29, Issue 12
The expressions of Caveolin-2 mRNAs, natural antisense transcripts and proteins in mouse liver development
Mitsuru Chiba*
Department of Bioscience and Laboratory Medicine, Graduate School of Health Sciences, Hirosaki University, 66-1 Hon-Cho, Hirosaki, Aomori, Japan
- *Corresponding Author:
- Mitsuru Chiba
Department of Bioscience and Laboratory Medicine
Graduate School of Health Sciences
Hirosaki University
66-1 Hon-Cho, Hirosaki, Aomori, Japan
Accepted date: May 23, 2018
DOI: 10.4066/biomedicalresearch.29-18-626
Visit for more related articles at Biomedical ResearchAbstract
Caveolin-2 (Cav2) has been shown to be a major component of the inner surface of caveolae and be involved in lipid metabolism, development and cancer. Natural Antisense Transcripts (NATs) are transcribed from a large number of genes in various species, including humans and mice. In the present study, we investigated the expressions and localization of Cav2 mRNAs, NATs and proteins in the mouse liver at various developmental stages. The expressions of Cav2 mRNAs and proteins in the liver were higher at the new-born stage (NB) than at days 14, 17, and 19 of the embryonic stage (E) and at day 3 after birth. The expressions of Cav2 NATs were lower than that of Cav2 mRNAs at each stage. On tissues, the expressions of Cav2 mRNAs and NATs were uniformly low in the liver at E14. The expressions of Cav2 mRNAs were predominantly strong in the hepatocytes at NB, whereas the expressions of Cav2 NATs were low in the hepatocytes at NB but strongly detected in specific cells. In conclusions, Cav2 mRNAs and NATs did not co-localized in the liver at NB. These results suggest that Cav2 NATs may be not involved in the regulation of Cav2 mRNA expressions at NB.
Keywords
Caveolin-2, Natural antisense transcripts, Liver development, In situ hybridization, New-born.
Introduction
Caveolae are specialized domains of the plasma membrane that are implicated in the sequestration of a variety of lipid and protein molecules [1]. It has been suggested that these important cellular organelles play a pivotal role in biochemical processes such as lipid metabolism, growth regulation, signal transduction, and apoptosis [2]. Caveolin interacts with and regulates heterotrimeric G-proteins [1,2]. Currently, there are 3 members of the caveolin multi-gene family that encode 21-24 kDa integral membrane proteins that comprise the major structural component of the caveolar membrane [3]. Caveolin-2 (Cav2) is abundantly expressed in fibroblasts, differentiated adipocytes, smooth and skeletal muscles, and endothelial cells [4]. The expressions of Cav1 are similar to that of Cav2, whereas that of Cav3 are limited to the muscle tissues [5]. Cav1 regulates hepatic lipid accumulation, lipid and glucose metabolism, and hepatic proliferation [6]; however, the expressions and functions of Cav2 in the liver are unknown.
Natural antisense transcripts (NATs) are known as one of gene expression regulators in eukaryotes [7]. In the past several years, the expression of NATs in mammals is reported [8,9]. In addition, investigation that the differential expressions of several NATs are induced during the process of liver development is made [10]. Due to the increase in the number of these studies, researchers have initiated investigations of not only the expressions of mRNAs and NATs but also the localization and regulation of their transcripts to completely understand the molecular interactions occurring in individual loci, cells, and tissues. Changes in the level of expressions of each NAT have been described in complex diseases such as cancer and neurological [11,12]. Identification of Cav2 NATs as an up-regulated gene during liver development is done [10]. However, little is known about the expressions of Cav2 mRNAs and NATs during pre and postnatal development of the liver. Hence, in the present study, the possible existence of Cav2 NATs and the localization of Cav2 mRNAs and its NATs during the development of the liver is examined.
Materials and Methods
Liver samples
Livers at various developmental stages, embryonic stage (E) days 14, 17, and 19, new-born stage (NB), and 3-d-old stage (3D), were obtained from the C57BL/6J mice at the RIKEN BioResource Center (Tsukuba, Ibaraki, Japan). All animal experiments were carried out according to the RIKEN guidelines for the care and use of experimental animals. Livers at E14, E17, E19, NB, and 3D were collected (each n=3).
Western blotting
Liver samples were lysed using M-PER Mammalian Protein Extraction Reagent (ThermoFisher Scientific, Inc., Waltham, MA, USA) supplemented with Halt proteinase inhibitor cocktail (ThermoFisher Scientific, Inc.). Protein concentrations were determined using the BCA Protein Assay Kit (ThermoFisher Scientific, Inc.) and Benchmark Microplate Reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA) according to the manufacturer’s instructions. Protein samples (10 μg) from livers at various developmental stages were prepared using 4X sample buffer (0.25 mol/l Tris-HCl, 8% SDS, 40% glycerol, 20% 2-mercaptoethanol and 0.02% bromophenol blue; pH 6.8). Electrophoresis and immunoblotting were performed by protocol reported previously [13]. Anti-Cav2 rabbit polyclonal antibody (#SP5143P, Acris Antibodies, Herford, Germany) (1:1000 dilution) and anti-β-actin (Actb) rabbit polyclonal antibody (#ab8227; Abcam, Cambridge, UK) (1:1000 dilution) were used as primary antibodies.
Strand-specific RT-qPCR
Total RNAs from the livers was isolated using Isogen II (Nippongene, Tokyo, Japan) according to the manufacturer’s instructions. The quality and concentration of the total RNAs were assessed using the NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) according to the manufacturer’s instructions. All the total RNA samples had 260/280 nm absorbance ratios of 1.8-2.0 [9]. To provide an external control for the comparison of results obtained from strand-specific reverse transcription-quantitative polymerase chain reaction (strand-specific RT-qPCR), an aliquot of each RNA sample was mixed with that of the RNA fragment synthesized from pEGFP-C1 vector (EGFP: enhanced green fluorescent protein; ThermoFisher Scientific, Inc.) to attain a final amount of 5 × 10-5 pmol/10 μg total RNA, and the resulting mixture was subjected to the synthesis of first-strand complimentary DNA (cDNA) [14].
Strand-specific RT-qPCR was performed using the total RNAs obtained from livers at different developmental stages to confirm the expression pattern of Cav2 mRNAs and NATs during liver development. First-strand cDNAs derived from mRNAs or NATs were synthesized using a Cav2 reverse primer (5’-ACA AAA CAA CTA CAT GTC TAC ATG-3’) or Cav2 forward primer (5’-TTT GGA ATG CAG TGT AAA ATGTG-3’), EGFP reverse primer (5’-GAA CTC CAG CAG GAC CATGT-3’), and reverse transcriptase (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The mixtures were incubated at 50°C for 60 min. The resulting cDNA was incubated with RNase A at 99°C for 5 min and at 37°C for 60 min to digest the RNAs. qPCR of mRNAs and NATs was performed using the first-strand cDNAs, FastStart Universal SYBR-Green Master (Roche Diagnostics, Basel, Switzerland) and primer pairs. qPCR analyses were performed by protocol reported previously [9] using the StepOne Plus Real-Time PCR system (ThermoFisher Scientific, Inc.). To determine quantity of RNA copies, standard curve method using EGFP expression was performed. The results of qPCR are presented as means ± standard errors of the samples.
In situ hybridization
To examine the expressions of Cav2 mRNAs and NATs on the liver, 120-nucleotide probes with a specific sequence were designed from the mRNAs of Cav2 (5’-UUU GGA AUG CAG UGU AAA AUG UGC UGU CUA GGU GUU CUC UCC AAC UUA CCC AGC AAA UGU UUU AUG ACU GGG CUU CAU CUG GCU GUG ACC AUA AGA CAU GUA GAC AUG UAG UUG UUU UGU-3’). As a negative-control RNA probe (5’-UGA CGG ACA UCG GGA AAC GCC AAA GGA GAU UAU GUA CCG AGG AAG AAU GUC GCU GGA CGG UAU GCA GGA AAA GGA GGA CGU GUG GCG AGA CAG CGA CGA AGU AUC ACC GAC AUA AUC UGC-3’), a 120-nucleotide λ-phage sequence, which had no similarity with any of the mammalian sequences registered in the DNA Data Bank of Japan (www.ddbj.nig.ac.jp/indexj. html), was used in all in situ hybridization experiments to verify that the hybridization system did not emit any nonspecific hybridization signals. Digoxigenin (DIG)-labeled cRNA probes were provided by Tsukuba GeneTech Laboratories (Ibaraki, Japan).
C57BL/6J mice at different developmental stages (E14 and NB) were obtained from the RIKEN BioResource Center (Ibaraki, Japan). For in situ hybridization, tissues from mice at different developmental stages were first fixed in situ by perfusion with 4% (w/v) ice-cold paraformaldehyde solution in phosphate-buffered saline (PBS). The resulting tissues were excised and further fixed overnight in paraformaldehyde solution. The fixed tissues were embedded in paraffin and cut into 4-μm sections. These sections were placed on glass slides and subjected to hematoxylin and eosin (HE) staining and in situ hybridization. In situ hybridization was performed using the de-paraffinized sections of embryos as described above. Hybridization, washing and detection were performed by protocol reported previously [15].
Results and Discussion
I have discovered that Cav2 NATs expressed in the liver and changed during liver development and liver regeneration using microarray analysis, previously [10,16]. However, the localization and functions of Cav2 NATs in the liver have been unknown until now. Therefore, the expressions of Cav2 mRNAs, NATs and proteins in the liver at various developmental stages were analyzed in the present study.
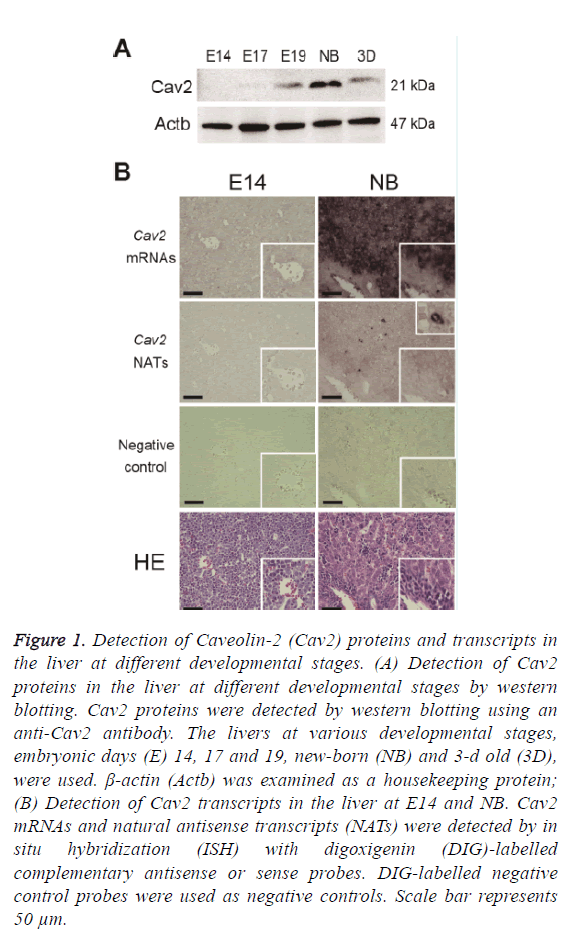
Livers from C57BL/6J mice at E14, E17, E19, NB, and 3D were used for the preparation of Cav2 protein expressions. Western blotting was performed as described in the Materials and Methods section. The expressions of Cav2 proteins were high in the liver at NB and low at E14 (Figure 1A). This suggests that Cav2 proteins may have important roles in the liver at NB. Liver at foetus works mainly as a hematopoietic organ. Cav2 proteins may be involved in a part of functional exchanges in the liver.
Figure 1: Detection of Caveolin-2 (Cav2) proteins and transcripts in the liver at different developmental stages. (A) Detection of Cav2 proteins in the liver at different developmental stages by western blotting. Cav2 proteins were detected by western blotting using an anti-Cav2 antibody. The livers at various developmental stages, embryonic days (E) 14, 17 and 19, new-born (NB) and 3-d old (3D), were used. β-actin (Actb) was examined as a housekeeping protein; (B) Detection of Cav2 transcripts in the liver at E14 and NB. Cav2 mRNAs and natural antisense transcripts (NATs) were detected by in situ hybridization (ISH) with digoxigenin (DIG)-labelled complementary antisense or sense probes. DIG-labelled negative control probes were used as negative controls. Scale bar represents 50 μm.
In strand-specific RT-qPCR, we showed that the amounts of Cav2 mRNAs in the liver at NB was 130.779 ± 75.505 × 10-5 pmol/10 μg total RNAs and was the highest among the examined liver development stages (Table 1). This result is consistent with the expressions of Cav2 proteins, as described in Figure 1A. There were no significant differences between the amounts of Cav2 NATs among the examined livers, whereas the expressions of Cav2 NATs were less than that of Cav2 mRNAs at each stage (Table 1). This indicates that the expressions of Cav2 NATs are low in liver during developmental process.
| Stage | Amount of transcripts per 10 mg total RNA | |
|---|---|---|
| Cav2 mRNAs | Cav2 NATs | |
| (Ã 10-5 pmol) | (Ã 10-5 pmol) | |
| Mean ± SEM | Mean ± SEM | |
| E14 | 39.813 ± 22.986 | 3.067 ± 1.771 |
| E17 | 66.598 ± 38.450 | 5.176 ± 2.988 |
| E19 | 73.688 ± 42.544 | 2.433 ± 1.404 |
| NB | 130.779 ± 75.505 | 3.977 ± 2.296 |
| 3D | 109.211 ± 63.053 | 3.672 ± 2.120 |
Table 1: Amounts of Cav2 mRNAs and NATs in the liver at different developmental stages.
The localization of Cav2 mRNAs and NATs in the liver at NB with high mRNA expressions and at E14 with low mRNA expressions was examined by in situ hybridization (Figure 1B). High expressions of Cav2 mRNAs were observed in the hepatocytes at NB than that at E14. Uniform localization of Cav2 mRNAs and NATs were observed in the liver at E14. Low expressions of Cav2 mRNAs were observed in the hematopoietic cells of the liver at NB; furthermore, low expressions of Cav2 NATs were observed in the hepatocytes at NB, but the expressions of Cav2 NATs were strongly detected in specific cells. High expressions of Cav2 mRNAs were observed in the hepatocytes at NB. Moreover, no colocalization of Cav2 mRNAs and NATs were observed in the liver at NB. Taken together, these results suggest that Cav2 NATs may not be directly involved in the regulation of mRNA expressions at NB. In the previous study, it has been reported that Ncam1 mRNAs and NATs co-localized in the Purkinje cells of the cerebellum [17]. Therefore, it is suggested that the functions of Cav2 NATs are different from that of Ncam1 NATs. Cav2 mRNAs and NATs may be co-localized in organs other than the liver and be involved in the regulation of gene expressions. The expressions of Cav2 mRNAs and NATs in other organs should be examined in future study.
In conclusion, the expressions of Cav2 NATs were lower than that of Cav2 mRNAs and were not co-localized with the Cav2 mRNAs. Therefore, Cav2 NATs may not regulate Cav2 mRNA expression. In future studies, the cells expressing Cav2 NATs should be identified to obtain novel information about these NATs.
References
- Gu X, Reagan AM, McClellan ME, Elliott MH. Caveolins and caveolae in ocular physiology and pathophysiology. Prog Retin Eye Res 2017; 56: 84-106.
- Gupta R, Toufaily C, Annabi B. Caveolin and cavin family members: dual roles in cancer. Biochimie 2014; 107: 188-202.
- Williams TM, Lisanti MP. The Caveolin genes: from cell biology to medicine. Ann Med 2004; 36: 584-595.
- Zhu LL, Cui Y, Chang YS, Fang FD. A second protein marker of caveolae: caveolin-2. Chin Med Sci J 2010; 25: 119-124.
- Galbiati F, Engelman JA, Volonte D, Zhang XL, Minetti C, Li M, Hou H, Kneitz B, Edelmann W, Lisanti MP. Caveolin-3 null mice show a loss of caveolae, changes in the microdomain distribution of the dystrophin-glycoprotein complex, and t-tubule abnormalities. J Biol Chem 2001; 276: 21425-21433.
- Fernandez-Rojo MA, Ramm GA. Caveolin-1 function in liver physiology and disease. Trends Mol Med 2016; 22: 889-904.
- Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 2009; 10: 637-643.
- Chiba M, Kimura M, Asari S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol Rep 2012; 28: 1551-1558.
- Chiba M, Miura T, Kasai K, Monzen S, Kashiwakura I, Yasue H, Nakamura T. Identification of up-regulated and down-regulated cis-natural antisense transcripts in the human B lymphoblastic cell line IM-9 after X-ray irradiation. Mol Med Rep 2012; 5: 1151-1157.
- Chiba M. Differential expression of natural antisense transcripts during liver development in embryonic mice. Biomed Rep 2014; 2: 918-922.
- Kohno K, Chiba M, Murata S, Pak S, Nagai K, Yamamoto M, Yanagisawa K, Kobayashi A, Yasue H, Ohkohchi N. Identification of natural antisense transcripts involved in human colorectal cancer development. Int J Oncol 2010; 37: 1425-1432.
- Guo JH, Cheng HP, Yu L, Zhao S. Natural antisense transcripts of Alzheimer's disease associated genes. DNA Seq 2006; 17: 170-173.
- Chiba M, Watanabe N, Watanabe M, Sakamoto M, Sato A, Fujisaki M, Kubota S, Monzen S, Maruyama A, Nanashima N, Kashiwakura I, Nakamura T. Exosomes derived from SW480 colorectal cancer cells promote cell migration in HepG2 hepatocellular cancer cells via the mitogen-activated protein kinase pathway. Int J Oncol 2016; 48: 305-312.
- Kiuchi S, Yamada T, Kiyokawa N, Saito T, Fujimoto J, Yasue H. Genomic structure of swine taste receptor family 1 member 3, TAS1R3, and its expression in tissues. Cytogenet Genome Res 2006; 115: 51-61.
- Chiba M, Kiyosawa H, Hiraiwa N, Ohkohchi N, Yasue H. Existence of Pink1 antisense RNAs in mouse and their localization. Cytogenet Genome Res 2009; 126: 259-270.
- Chiba M, Murata S, Myronovych A, Kohno K, Hiraiwa N, Nishibori M, Yasue H, Ohkohchi N. Elevation and characteristics of Rab30 and S100a8/S100a9 expression in an early phase of liver regeneration in the mouse. Int J Mol Med 2011; 27:567-574.
- Chiba M. Detection of natural antisense non-coding RNAs transcribed from Ncam1 in mice tissues at several developmental stages. Biomed Rep 2015; 3: 663-667.