Review Article - Journal of RNA and Genomics (2022) Volume 18, Issue 2
The "escape" of transposons in drosophila models of central nervous system diseases: An integrated overview.
Benedetta Saccomanno, Valeria Specchia*
Department of Biological and Environmental Sciences and Technologies, University of Salento, Lecce LE, Italy
- Corresponding Author:
- Valeria Specchia
Department of Biological and Environmental Sciences and Technologies, University of Salento, Lecce LE, Italy
E-mail: valeria.specchia@unisalento.it
Abstract
Transposable elements are repetitive sequences widely present in eukaryotic genomes, embedded in the heterochromatin-the tightly condensed chromatin-which prevents their transposition. The unscheduled transcription of transposons and its harmful consequences have been proven to play a role in neuron degeneration. Drosophila models are crucial to unveiling this role and the mechanisms triggering transposon activity. Specifically, abnormal heterochromatin relaxation, which is also observed in Alzheimer’s disease, has been found as the key event leading to transposon “escape” in the brain. This review recapitulates the main efforts towards demonstrating the role of transposons in neurodegeneration focusing on drosophila models, and offers an integrated overview of common and specific molecular mechanisms useful for identifying new therapeutic targets.
Keywords
Drosophila, Transposons, Neurodegeneration.
The Structural and Mechanistic Features of Transposons in Drosophila
The heterochromatin of Drosophila melanogaster corresponds to one third of the whole genome and it is concentrated in the telomeric and pericentromeric regions [1,2]. The Transposable Elements (TEs) are known to be major structural components of drosophila heterochromatin, which also includes other peculiar repetitive sequences, such as stellate and suppressor of stellate, as well as some coding genes and satellite repeats [3-5]. More recently, assembly and mapping of the repetitive sequences of drosophila heterochromatin were generated. In addition, complete sequencing of euchromatic genomic regions of Drosophila melanogaster has provided the sequence and insertion sites for the repetitive elements in the drosophila euchromatin [6-9].
The embedding of transposable elements into heterochromatin ensures their transcriptional silencing and avoids their movement [10]. Transposable elements are mainly repressed by H3K9 di/tri-methylation, the epigenetic hallmark for HP1 (Heterochromatin Protein 1). SU (VAR) 3–9 encodes a histone methyl transferase which selectively methylates histone H3 at lysine 9 (H3K9) ensuring HP1 binding, spreading and chromatin condensation [11,12]. In fact, the constitutive heterochromatin or “green chromatin” that is marked by SU (VAR) 3-9, HP1, and enriched of H3K9me2 histone mark, contains closely packed DNA that is transcriptionally repressed. Mapping studies revealed multiple binding of HP1 to TEs [13-15].
In addition to transcriptional silencing, transposable elements undergo a post-transcriptional silencing mediated by Argonaute proteins and specific classes of small non coding RNAs [16]. In drosophila germ cells, the Argonaute proteins Ago3, piwi and aubergine and the small non coding RNAs piRNAs are the key players in the post-transcriptional transposons silencing [17]. In drosophila somatic cells, including neurons, Argonaute protein Ago2 and ribonuclease Dicer2, in concert with the small RNAs endo siRNAs, silence transposable elements by degrading their transcripts [18].
The unscheduled activity of transposons has dangerous effects for the cell as a result of different genome alterations. The new insertion of transposons can cause not only gene mutations, but also changes in global transcript levels in cells, by providing promoters or altering chromatin state [19].
Studies conducted in drosophila unveiled a de-regulation of transposons due to gene mutations responsible of human nervous system diseases. This review offers an integrated vision of these recent studies related to transposons activation in altered conditions of the brain [20].
Transposable Elements are Dysregulated in Heads of Transgenic Drosophila Expressing Human Mutant Tau
Alzheimer’s disease is an age-related degenerative neurological disorder that affects the brain [21]. The main hallmarks identifying Alzheimer’s brain include the extracellular presence of aggregates of amyloid. A peptide and the tau neurofibrillary tangles in neuronal cell bodies or neuritis. Tau is a microtubule-associated protein, which stabilizes neuronal microtubules and, thus, ensures their functions. In Alzheimer’s disease tau is abnormally hyper-phosphorylated, affecting its ability to bind microtubules which consequently disassemble [22-24].
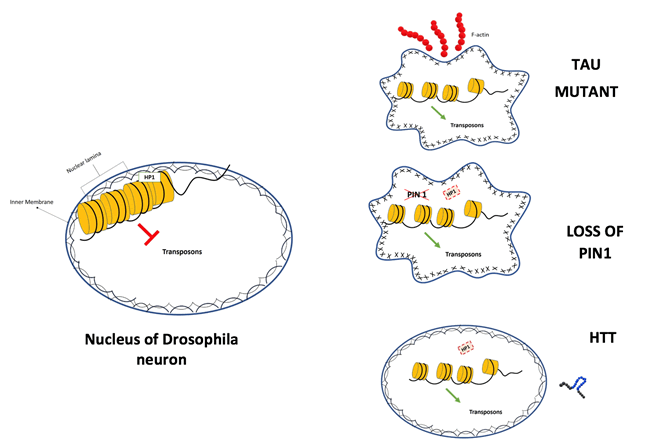
Transgenic drosophila, expressing mutant form of human tau, shows age-related and progressive neurodegeneration in specific brain regions without the accumulation of neurofibrillary tangles. RNAseq and qPCR analyses revealed the activation of multiple transposons in human tau transgenic drosophila heads. The dysregulation of transposable elements has been related to heterochromatin decondensation. In addition, the Piwi protein and piRNAs depletions were observed in tau transgenic drosophila, identifying a Piwi-mediated mechanism causing transposons activation. Double stranded RNAs derived from transcriptional release of transposons contribute to an inflammation state in neurons.
The evidence from drosophila models is in agreement with TE activation highlighted in brains of tau transgenic mice and in brains from deceased individuals with Alzheimer’s disease. In tau transgenic drosophila, heterochromatin relaxation is linked to nuclear lamin dysfunction. Mechanistically, Tau stabilizes the actin filaments that in turn cause the reduction and disorganization of the nuclear lamin in neurons [25].
Pin1-Mediated Mechanism Underlies Heterochromatin Maintenance and Transposons Control in Drosophila Brain
In the cell nucleus, heterochromatin is mainly located at the nuclear periphery and it is tightly associated to the nuclear lamin in the LADs (Lamina-Associated Domains).
The nuclear lamin is a network of A- and B-type lamins, intermediate filamentous proteins coating the inner nuclear membrane, that interact with multiple proteins such as Barrier to Autointegration Factor (BAF), LAP proteins, Lamin B Receptor (LBR) or Emerin. Proteins of the LINC complex such as Nesprins and Sun proteins connect nuclear lamin and cytoskeletal components across the nuclear envelope. Nuclear lamin has a crucial role in maintaining chromatin organization, and nucleus shape under mechanical intra- and extracellular forces. Cells from individuals with a Lamin A mutation present a loss of heterochromatin, causing Hutchinson Gilford Progeria Syndrome [26-28].
Studies in drosophila, confirmed in mice and human, have provided advancement towards understanding the molecular mechanisms underlying the relationship between mechanical cues, nuclear lamina and heterochromatin organization, by attributing a key role to the prolyl-isomerase Pin1.
Mechanistically, in the fly brain Pin1 maintains lamin B structure in a phosphorylation-dependent manner ensuring HP1a binding and stability. Loss of Pin1 decreases HP1a stability and causes heterochromatin relaxation and transposons dysregulation. These experiments demonstrated the fundamental role of Pin1 in controlling transposons, preserving neuron survival and cognitive functions [29]. The action of Pin1 is specifically essential during mechanical stress, preventing nuclear shape deformation and heterochromatin relaxation in these stressful conditions. Interestingly, the reduction of Pin1 takes place in Alzheimer affected brains and has an effect in the maintenance of the nuclear envelope structure and heterochromatin condensation [30,31].
Transposable Elements are Dysregulated in Drosophila Model of Huntington Disease
Huntington Disease (HD) patients present an array of motor, cognitive and behavioural deficits. The disease is caused by the expansion of a CAG trinucleotide repeat in HTT gene that encodes the protein huntingtin. The mutant huntingtin protein has a long polyglutamine sequence and results toxic to neurons. The feature of the disease is a progressive loss of spiny neurons of the striatum [32,33].
In larval and adult brains from transgenic drosophila expressing human mutant huntingtin, the global heterochromatin relaxation drives the deregulation of transposons. Regarding the transposable element GYPSY_I, the increase of RNA expression level correlates with a higher number of genomic insertion sites in HD fly brains than in controls [34]. However, other studies need to be performed to establish the potential role of transposons in huntington disease. In a recent study, the RNA and protein levels of the transposon Line1 have been investigated in a mouse model of HD. The Line1 RNA expression level decreases in HD, but the expression of Orf1 and Orf2 doesn’t correlate with RNA levels and it increases in specific regions of the brain and at specific age [35-39].
Discussion
Unscheduled transcription of transposons causes harmful effects in neurons, leading to neurodegeneration. Interestingly, the use of inhibitors of the retro-transcriptase in drosophila models had rescued neurodegeneration effects from cellular to phenotypic level. The relaxation of the heterochromatin structure appears the principal and common event at the base of transposon transcriptional release (Figure 1). However, the mechanisms that lead to chromatin decondensation are different as revised in this review [40-43].
Conclusion
The “transposons” escape in neurons is emerging as a mechanism that contributes to neurodegenerative processes. Therefore, the genetic and molecular dissection of the mechanisms underlying the transposon activation in neurons and in age-dependent manner is crucial to unlock therapeutic targets.
References
- Bates GP, Dorsey R, Gusella JF, et al. Huntington disease. Nat Rev Dis Primers. 2015;1:15005.
[Crossref], [Google Scholar], [Indexed]
- Casale MA, Liguori F, Ansaloni F, et al. Transposable element activation promotes neurodegeneration in a drosophila model of huntington's disease. iScience. 2022; 25(1):103702.
[Crossref], [Google Scholar], [Indexed]
- Chénais B, Caruso A, Hiard S, et al. The impact of transposable elements on eukaryotic genomes: From genome size increase to genetic adaptation to stressful environments. 2012;509:715.
[Crossref], [Google Scholar], [Indexed]
- Cohen CJ, Lock WM, Mager DL. Endogenous retroviral LTRs as promoters for human genes: A critical assessment. 2009;448:105–114.
[Crossref], [Google Scholar], [Indexed]
- Bourque G, Leong B, Vega VB, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–1762.
[Crossref], [Google Scholar], [Indexed]
- Eissenberg JC, James TC, Hartnett DMF, et al. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc Natl Acad Sci USA. 1990;87:9923-9927.
[Crossref], [Google Scholar], [Indexed]
- Filion GJ, van Bemmel JG, Braunschweig U et al. Systematic protein location mapping reveals five principal chromatin types in drosophila cells. Cell. 2010;143(2):212-224.
[Crossref], [Google Scholar], [Indexed]
- Floreani L, Ansaloni F, Mangoni D, et al. Analysis of LINE1 retrotransposons in Huntington's disease. Front Cell Neurosci. 2022;15:743797.
[Crossref], [Google Scholar]
- Frost B, Hernberg M, Lewis J, et al. Tau promotes neurodegeneration through global chromatin realaxation. Nature neuroscience. 2022;17:357-366.
[Crossref], [Google Scholar], [Indexed]
- Bozzetti MP, Fanti L, Di Tommaso S, et al. The "Special" crystal-Stellate system in Drosophila melanogaster reveals mechanisms underlying piRNA pathway-mediated canalization. Genet Res Int. 2012;324293.
[Crossref], [Google Scholar]
- Ghildiyal M, Seitz H, Horwich D, et al. Endogenous siRNAs derives from transposons and mRNAs in drosophila somatic cells. Science. 2008;320(5879):1077-1081.
[Crossref] [Google Scholar] [Indexed]
- Goldman RD, Shumaker DK, Erdos MR, et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson–Gilford progeria syndrome. Proc Natl Acad Sci USA. 2004;101(24):8963-8968.
[Crossref], [Google Scholar], [Indexed]
- Chang CH, Larracuente AM. Heterochromatin-enriched assemblies reveal the sequence and organization of the Drosophila melanogaster Y chromosome. Genet. 2019;211(1):333-348.
- Greil F, van der Kraan J, Delrow J, et al. Distinct HP1 and Su(var)3-9 complexes bind to sets of developmentally coexpressed genes depending on chromosomal location. Gens Dev. 2003;17(22):2825-2838.
[Crossref], [Google Scholar]
- Guo C, Jeong HH, Hsieh YC, et al. 2018. Tau activates transposable elements in alzheimer's disease. Cell Rep. 2018;23:2874–2880.
[Crossref], [Google Scholar], [Indexed]
- Hoskins RA, Carlson JW, Kennedy C, et al. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science. 2007;316:1625-1628.
[Crossref], [Google Scholar], [Indexed]
- Drewes G, Ebneth A, Mandelkow EM. MAPs, MARKs and microtubule dynamics. Trends Biochem Sci. 1998;23:307-311.
[Crossref], [Google Scholar], [Indexed]
- James TC, Elgin SC. Identification of a non-histone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol. 1986;6(11):3862-3872.
[Crossref], [Google Scholar], [Indexed]
- Kirby TJ, Lammerding J. Emerging views of the nucleus as a cellular mechanosensor. Nat Cell Biol. 2018;20:373-381.
[Crossref], [Google Scholar], [Indexed]
- Al Bassam J, Ozer RS, Safer D, et al. MAP2 and tau bind longitudinally along the outer ridges of microtubule protofilaments. J Cell Biol. 2002;157(1):1187-1196.
[Crossref], [Google Scholar], [Indexed]
- Eissenberg JC, Elgin S. The HP1 protein family: getting a grip on chromatin. Curr Opin Genet Dev. 2000;10(2):204-210.
[Crossref], [Google Scholar], [Indexed]
- Kris ND, Ribeiro AJS, Lammerding J. Nuclear shape, mechanics, and mechanotransduction. Circ Res. 2008;102:1307-1318.
[Crossref], [Google Scholar], [Indexed]
- Li W, Prazak L, Chatterjee N, et al. Activation of transposable elements during aging and neuronal decline in drosophila. Nat Neurosci. 2013;16:529-531.
[Crossref] [Google Scholar], [Indexed]
- Malone CD, Brenencke J, Dus M, et al. Specialized piRNA pathways act in germline and somatic tissues of the drosophila ovary. Cell. 2009;137(3):522-535.
[Crossref], [Google Scholar], [Indexed]
- Mandelkow EM, Mandelkow E. Tau in alzheimer's disease. Trends Cell Biol. 1998;8(11):425-427.
[Crossref], [Google Scholar], [Indexed]
- Pimpinelli S, Berlolo M, Fanti L, et al. Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Proc Natl Acad Sci USA. 1995;92(9):3804-3808.
[Crossref], [Google Scholar], [Indexed]
- Prokocimer M, Davidovich M, Rafinia MN, et al. Nuclear lamins: key regulators of nuclear structure and activities. J Cell Mol Med. 2009;13(6):1059-1085.
[Crossref], [Google Scholar], [Indexed]
- Ramirez P, Zuniga G, Sun W, et al. Pathogenic tau accelerates aging-associated activation of transposable elements in the mouse central nervous system. Prog Neurobiol. 2002;208:102181.
[Crossref], [Google Scholar] [Indexed]
- Napoletano F, Bravo GF, Voto IAP, et al. The prolyl-isomerase PIN1 is essential for nuclear Lamin-B structure and function and protects heterochromatin under mechanical stress. Cell Rep. 2021;36(11):109694.
[Crossref], [Google Scholar], [Indexed]
- Schotta G, Ebert A, Krauss V, et al. 2002. Central role of drosophila SU(VAR)3–9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121-1131.
[Crossref], [Google Scholar], [Indexed]
- Specchia V, D'Attis S, Puricella A, et al. dFmr1 Plays Roles in Small RNA Pathways of Drosophila melanogaster. Int J Mol Sci. 2017;18(5):1066.
[Crossref], [Google Scholar], [Indexed]
- Napoletano F, Bravo GF, Voto IAP, et al. The prolyl-isomerase PIN1 is essential for nuclear Lamin-B structure and function and protects heterochromatin under mechanical stress. Cell Rep. 2021;36(11):109694.
[Crossref], [Google Scholar], [Indexed]
- Spradling AC, Rubin GM. drosophila genome organization: conserved and dynamic aspects. AnnRev Genet. 1989;15:219-264.
[Crossref], [Google Scholar], [Indexed]
- Yamamoto M, Mitchelson A, Tudor M, et al. 1990. Molecular and cytogenetic analysis of the heterochromatin-euchromatin junction region of the Drosophila melanogaster X chromosome using cloned DNA sequences. Genetics. 1990;125:821-832.
[Crossref], [Google Scholar], [Indexed]
- Wang J, Keightley PD, Halligan DL. Effect of divergence time and recombination rate on molecular evolution of drosophila INE-1 transposable elements and other candidates for neutrally evolving sites. J Mol Evol. 2007;65:627.
[Crossref], [Google Scholar], [Indexed]
- Wittmann CW, Wszolek MF, Shulman JM, et al. Tauopathy in drosophila: neurodegeneration without neurofibrillary tangles. Science. 2001;293(5530):711-714.
[Crossref], [Google Scholar], [Indexed]
- Vonsattel JP, Difiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57(5):369-384.
[Crossref], [Google Scholar], [Indexed]
- Thomas EO, Ramirez P, Hyman BT, et al. 2021. Testing the neuroinflammatory role of tau-induced transposable elements in tauopathy. Basic Science. 2021;17(2):e058664.
[Crossref], [Google Scholar]
- van Steensel B, Belmont AS. Lamina-associated domains: links with chromosome architecture, heterochromatin and gene repression. Cell. 2017;169(5):780-791.
[Crossref], [Google Scholar], [Indexed]
- Sun W, Samimi H, Gamez M, et al. Pathogenic tau-induced piRNA depletion promotes neuronal death through transposable element dysregulation in neurodegenerative tauopathies. Nat Neurosci. 2021;21:1038-1048.
[Crossref], [Google Scholar], [Indexed]
- van Steensel B, Delrow J, Henikoff S. Chromatin profiling using targeted DNA adenine methyltransferase. Nat Genet. 2001;27(3):304-308.
[Crossref], [Google Scholar], [Indexed]
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318(5851):761-764.
[Crossref], [Google Scholar], [Indexed]
- Adams MD, Celniker SE, Holt RA, et al. The genome sequence of drosophila melanogaster. Science. 2000;287(5461):2185-2195.
[Crossref], [Google Scholar], [Indexed]