Research Article - Biomedical Research (2017) Volume 28, Issue 20
Role of microRNA-18a in the development of inner ear
Liang Sun1,2#, Chang Dong2#, Xianren Wang1, Kaitian Chen1, Changwu Li2, Geng Xu1* and Hongyan Jiang2*
1Department of Otorhinolaryngology, the First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, Guangdong, PR China
2Department of Otorhinolaryngology, Hainan Provincial People's Hospital, Haikou, Hainan, PR China
#These authors contributed equally
- *Corresponding Author:
- Hongyan Jiang
Department of Otorhinolaryngology
Hainan Provincial People's Hospital, PR China
Accepted date: November 03, 2017
Abstract
Objective: The developmental process of vertebrate inner ear is complex. MicroRNAs (miRNAs) have been reported to be involved in the development of inner ear. The current study was designed to explore the potential role of miR-18a in the development of inner ear.
Methods: The expression of miR-18a and lysine demethylase 5B (Kd5mb) in inner ear tissues and house Ear Institute-Organ of Corti 1 (HEIOC1) cells was detected by miRNA in situ hybridization (MISH) and immunofluorescence (IF), respectively. TargetScane was used to predict the potential target genes of miR-18a. Luciferase reporter assay, real-time PCR, and Western blot were performed to confirm that Kdm5b is a target gene of miR-18a. In addition, the effects of miR-18a aberrant expression on H3K4me3 level were detected by Western blot.
Results: The results showed that the expression of miR-18a was gradually increased and the expression of Kdm5b was gradually decreased during the inner ear development. Kdm5b was confirmed as a target gene of miR-18a and was negatively regulated by miR-18a. Moreover, the results showed that the expression of H3K4me3 was enhanced by miR-18a mimic but weakened by miR-18a inhibitor.
Conclusion: This study provides evidence that miR-18a and Kdm5b was dynamically regulated in the developmental process of inner ear. The effects of miR-18a on the inner cell development might be by directly targeting Kdm5b and indirectly regulating H3K4me3.
Keywords
MiR-18a, Kdm5b, H3K4me3, Inner ear development
Introduction
Hearing loss is the most common sensory defect, which afflicts hundred million people worldwide [1]. The vertebrate inner ear is responsible for both hearing and balance. The development process of inner ear is very complex, requiring coordinated transformation of a uniform sheet of cells into a labyrinth with multiple cell types [2]. Generally, the original cause of hearing loss is associated with a loss of hair cells. Various destructive factors, such as aging, aminoglycosides, neomycin, reactive oxygen species (ROS) accumulation, ect [3-5], have been reported to be responsible for the loss of hair cells. Therefore, it is of significance to understand the pattern of gene expression regulating the process of development of the inner ear.
Although the underlying mechanisms have not been fully understood, one strong candidate is microRNA (miRNA). MicroRNAs (miRNAs) are small, non-coding RNAs that regulate gene expression at post-transcriptional level by base pairing to the 3'- untranslated regions (UTR) of their target mRNAs [6]. Increasing evidence suggests that miRNAs are involved in the development and function of inner ear hair cells [7-10]. For example, Sacheli et al. found that the expression of miR-96, miR-182 and miR-183 was highly dynamic during the development of the cochlea [9]. Among the miRNAs, the functions of miR-18a have been recently investigated. It has been suggested that miR-18a is involved in different cancers, which is responsible for the cell proliferation, apoptosis, and differentiation [11-13]. More recently, miR-18a has been indicated in the development and morphogenesis of zebrafish inner ear [7]. However, the exact mechanism needs to be further explored.
The histone lysine demethylase Kdm5b is a member of the ATrich DNA interaction domain (ARID) containing the JmjC family of demethylases that specifically targets histone H3 lysine 4 (H3K4) [14]. Previous studies confirmed that Kdm5b regulates gene transcription and cell differentiation [15,16] and it is also indispensable for embryonic stem cell self-renewal, cell fate, and murine embryonic development, and preimplantation embryo development [17]. Nevertheless, the functional role of Kdm5b in the development of inner ear and hair cells is unclear.
Therefore, in the present study, we aimed to explore the functional roles of miR-18a and Kdm5b in the development of inner ear, as well as the underlying regulatory mechanism. Our study might provide a new insight into a better understanding of the role of miRNAs in the development of inner ear.
Materials and Methods
Cell lines and cell culture
House Ear Institute-Organ of Corti 1 (HEI-OC1) cells were obtained from Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University and University of Minnesota, Twin Cities. HEIOC1 cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco BRL, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and 50 U/mL interferon-γ without antibiotics at 33°C with an atmosphere of 10% CO2.
Animals
SPF C57/BL6 mice were purchased from Shanghai silaike experimental animal center. All the animals were kept in the First Affiliated Hospital of Zhongshan University animal center. The breeding pairs were placed together under a condition of on a 14:10 light–dark cycle and were allowed free access to food and water. The first 24 h following mating were considered as embryonic day 0 (E0) and the noon of the dates of the plugs were considered embryonic day 0.5 (E.5). Pregnant females were sacrificed under anesthesia and the embryos were dissected in ice-cold PBS at E 10–16, counting noon of the day was found as E10.5-E16.5. In addition, the first 24 h following birth were regarded as postnatal day 0 (P0). Embryos and new born mice (P0, P14, and P28) were fixed in 4% paraformaldehyde (PFA, Sigma-Aldrich, St Louis, MO) in 0.1 M phosphate buffer (pH 7.4). Animal experiments were approved by the First Affiliated Hospital of Zhongshan University ethics committee.
miRNA in situ hybridization (MISH)
The expression of miR-18a was detected by MISH, which was performed as previously described [18]. Inner ear tissues from the mice were fixed with 4% PFA. Inner ear sections (5-μm) deparaffinized, dehydrated and then fixed in 10% neutralbuffered formalin for 10 min. The sections were then immersed in acetylation solution and incubated in proteinase K for 15 min at 37°C. After digestion, the sections were immersed in RNase-free water and then they were prehybridized and hybridized with miRNA probe or control probe (0.5 μM). Subsequently, the samples were incubated at 37°C for 24 h, washed twice with standard saline citrate, and blocked in FBS and H2O2 at room temperature for 2 h. Staining was developed using nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP; Sigma-Aldrich).
Immunofluorescent (IF) staining
The immunofluorescent staining was performed as previously reported [19]. Briefly, the cochlear section or culture cells were fixed in 4% paraformaldehyde for 30 min, washed with phosphate buffer solution (PBS), and incubated in a blocking solution (10% goat serum and 1% bull serum albumin (BSA) in PBS) for 30 min with 0.1% Triton X-100. The cochlear section or culture cells were subsequently incubated with the following primary antibodies: monoclonal mouse anti-Kdm5b (1:50, ab56759, Abcam, Cambridge, MA, USA), and polyclonal rabbit anti-H3K4me3 (1:250, ab8580, Abcam) in the blocking solution at 4°C overnight. The following secondary antibodies were used: Alexa Fluor 488 conjugated goat anti-rabbit, Alexa Fluor 568 conjugated goat anti-mouse (Life Technologies). After being washed with PBS, the section or cells were incubated with corresponding Alexa Fluor 488- conjugated goat anti-mouse IgG or Alexa Fluor 568- conjugated goat anti-rabbit IgG (1:500, Molecular Probes) in the blocking solution at room temperature for 1 h. The sections or the cells were stained by 4’, 6-diamidino-2-phenylindole (DAPI, 0.1 mg/ml, D1306; Molecular Probes) for 20 min to visualize cell nuclei. After completely washing with PBS, the sections or the cells were mounted with a fluorescence mounting medium (H-1000, Vector Lab, CA) and were observed under a fluorescence microscope (Nickon, T2000) or a confocal microscope (Leica TCS SP2). The fluorescent image was saved in the TIFF format and assembled in Photoshop (Adobe Systems, CA) for presentation.
RNA extraction and quantitative real-time PCR
Total RNA was extracted from tissues or cell culture samples using TRIZOL reagent (Invitrogen) following the manufacturer’s protocol. The RNA was reverse transcribed into cDNA with a M-MLV reverse transcriptase kit (Promega, USA). Quantitative real-time PCR was performed using the Thermal Cycler Dice Detection System with the SYBR Premix Ex Taq™ (Takara, Japan). All samples were done in triplicate and the housekeeping gene β-actin was used as an endogenous control. The following primers were used to specially amplify the Kdm5b gene and β-actin. For Kdm5b, forward: 5’- GGTGAGCCAAAAACCTGGTA-3’, and reverse, 5’-AATCA CAAACTCCCCAGCAC-3’. For β-actin: forward: 5’- AGAGCCTCGCCTTTGCCG ATCC-3’, and reverse, 5’- CTGGGCCTCGTCGCCCACATA-3’. PCR conditions were listed as following: initial denaturation was performed at 95°C for 10 sec, followed by 35 cycles of denaturation at 96°C for 15 sec, annealing at 62?C for 5 sec, extension at 72°C for 40 sec, and then the final extension was at 72°C for 10 min. Gene expression was quantified using the 2-ΔΔCq method [20].
miRNA extraction and quantitative PCR measurement
miRNAs were extracted by mirVana™ miRNA Isolation Kit (AM1560, Ambion, USA) following manufacturer’s instructions. The purity and quantity of miRNA was determined by a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Inc., Rockland, DE). Then, miRNAs were converted into cDNA using TaqMan ® MicroRNA Reverse Transcription Kit (#4366596, Applied Biosystems, CA, USA) and measured by use of MyiQ real-time PCR detection system (Bio-Rad Laboratories) with TaqMan ® MicroRNA Assay (Applied Biosystems, CA, USA). An internal standard U6 snRNA was used as an internal control. The sequence of mRNA was listed as following: miR-18a, forward: 5’-UAAGGUGCAUCUAGUGCAGAUAG-3’, and reverse, 5’-AUCUGCACUAGAUGCACCUUAUU-3’; U6, Forward: 5’-CTCGC TTCGGCAGCACA-3’; Reverse: 5’- AACGCTTCACGAATTTGCGT-3’. The relative quantity of miRNA expression was calculated from the standard curve 39 and normalized to the amount of the internal standard U6.
Protein extraction and western blot analysis
Total protein was extracted using by using an extraction kit (Thermo Fisher Scientific, Waltham, MA, USA). Protein concentration was determined using a BCA Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.) according to the manufacturer’s instruction.. Protein samples (20 mg) were resolved using 10-15% SDS-PAGE and transferred onto a nitrocellulose membrane. The membrane was blocked with 5% milk in 0.1% Tris-buffered saline-Tween (TBST) for 2 h at room temperature. After incubation with the appropriate following primary antibodies overnight at 4 °C: anti-H3K4me3 (1:1000, ab8580, Abcam), anti-Kdm5b (1:1000, ab56759, Abcam) and anti-β-actin (1:2000, ab8226, Abcam), the membranes were washed and incubated with the IRDye 800cW or 680RD secondary antibodies in TBST for 2 h at room temperature. Then the protein bands of interest were visualized with the Odyssey system (LI-COR, USA).
miR-18a target gene prediction
According to the miRNA sequence provided by miRBase, the miRNA target gene databases TargetScan (http://www.targetscan.org/) were used for the prediction of the target genes of miR-18a.
Plasmid construction, cell culture and transfection
The reporter plasmid was constructed by inserting Kdm5b mRNA 3'UTR between XhoI and BamH I sites downstream of the Firefly luciferase gene in the psiCHECK-2 vector. 293T cells were cultured as above described and plated 24 h before transfection in 96-well plates at 2 × 104 cells/well. Cells were transfected with 0.2 μg psiCHECK-2-Kdm5b 3'UTR or psiCHECK-2, 0.45 μg pre-miRNAs or control and 0.15 μg sensor reporter DNA per well using Lipofectamine 2000 reagent (Life Technologies Inc.) according to the manufacturer’s instructions. All the experiments were performed in triplicate.
Dual-luciferase reporter assay
The full-length human 3’UTR of Kdm5b containing the putative binding site of miR-18a was amplified by PCR and cloned in pmirGLO Dual-Luciferase miRNA target expression vector (Promega; pmiR-3’UTRWT). This construct was used to generate plasmids carrying the mutated forms of Kdm5b 3’UTR, modified in the predicted miR-18a-binding sites (pmiR-3’UTRMut), using QuikChange II XLSite-Directed Mutagenesis Kit (Agilent). HEI-OC1 cells were seeded in 12- well plates and cotransfected with 500 ng pmiR-3’UTR (WT, Mut) and 100 nmol/L of either miR-18a precursor or negative control using Lipofectamine 3000. After 24 h of transfection, firefly and Renilla luciferase activities were measured by dual- Luciferase Reporter Assay System (Promega).
Statistical analysis
All data were presented as the mean ± standard deviation (SD). Statistical differences were compared by student's t-test or analysis of variance (ANOVA). The data were analyzed by using SPSS 17.0 (SPSS Inc., Chicago, IL, USA). P<0.05 was considered as significant difference between values.
Results
Expression levels of miR-18a in inner ear tissue
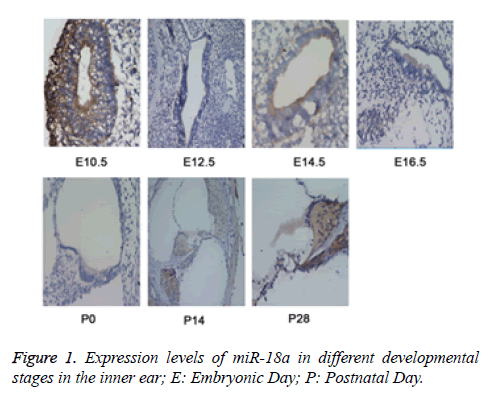
To explore the functional role of miR-18a in the development of inner ear, the MISH method was used to detect the expression of miR-18a at different time points (E10.5, E12.5, E14.5, E16.5, P0, P14, and P28) in the inner ear tissues from the mice. E10.5 represented as time for the development of acoustic vesicle; E12.5 represented as the development of vestibular sensory epithelium; E14.5 and E16.5: represented as the time for development of sensory epithelium; P0, P14, and P28 represented as the time for development of vestibule, corti and spiral neuron. The results revealed that during inner ear development, miR-18a was widely expressed not only in the cochlear hair cells and spiral ganglion but also in vestibular sensory cells (Figure 1), indicating that miR-18a may play an essential role in the inner ear development by maintenance of pluripotency of inner ear stem cells and differentiation of inner hair cells.
The expression levels of miR-18a were detected by miRNA in situ hybridization (MISH). The results showed that the levels of miR-18a were gradually increased during the inner ear development. E10.5 represented as time for the development of acoustic vesicle; E12.5 represented as the development of vestibular sensory epithelium; E14.5 and E16.5: represented as the time for development of sensory epithelium; P0, P14, and P28 represented as the time for development of vestibule, Corti and spiral neuron.
Expression levels of Kdm5b in inner ear tissue
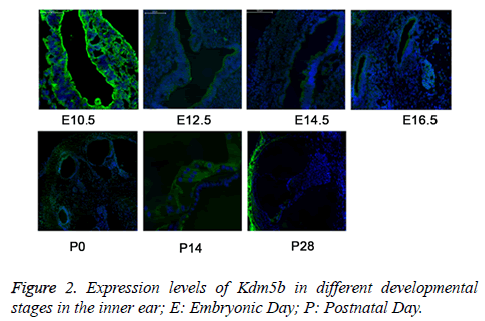
We then detected the expression of Kdm5b in the inner ear at the different time points (E10.5, E12.5, E14.5, E16.5, P0, P14, and P28) by the IF method. The results revealed that the expression of Kdm5b was gradually decreased during the inner ear development and maturation (Figure 2), indicating that Kdm5b was essential for the pluripotency maintenance of hair cell precursors but not for the differentiation regulation of cochlear hair cells.
The expression levels of Kdm5b were detected by immunofluorescence (IF). The results showed that the levels of Kdm5b were gradually decreased during the inner ear development. E10.5 represented as time for the development of acoustic vesicle; E12.5 represented as the development of vestibular sensory epithelium; E14.5 and E16.5: represented as the time for development of sensory epithelium; P0, P14, and P28 represented as the time for development of vestibule, corti and spiral neuron.
Expression levels of miR-18a and Kdm5b in HEI-OC1 cells
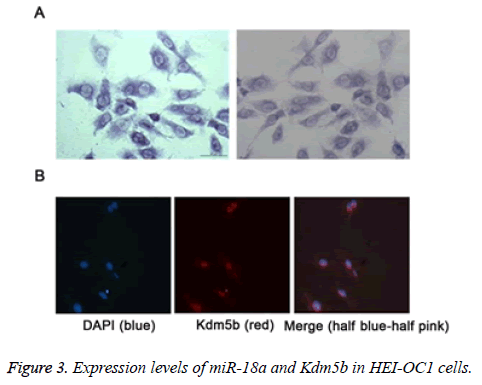
We further detected the expression levels of miR-18a and Kdm5b in the HEI-CO1 cells by MISH and IF, respectively. As shown in the Figures 3A and 3B, the results revealed that miR-18a and Kdm5b was expressed in the HEI-OC1 cells. The expression levels of miR-18a (A) and Kdm5b (B) in HEI-OC1 cells were respectively detected by miRNA in situ hybridization (MISH) and immunofluorescence (IF).
Kdm5b is a target gene of miR-18a
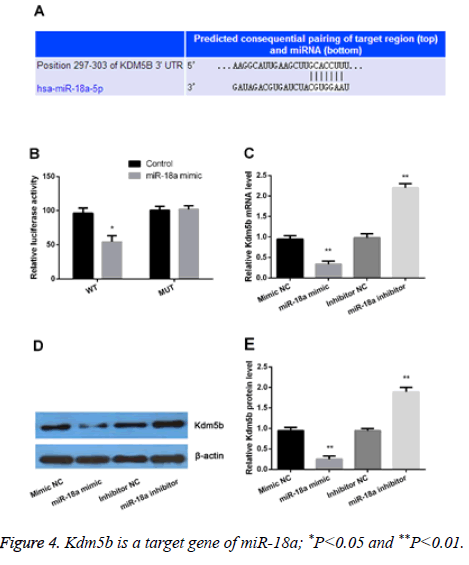
Kdm5b plays a critical role in cell development and proliferation. In the present study, our results showed that both the miR-18a and Kdm5b were differently expressed. Therefore, we assumed that Kdm5b might be a target gene of miR-18a. As shown in the Figure 4A, Kdm5b was predicated as a target gene of miR-18a by TargetScan. We further performed luciferase reporter assay, PCR and Western blot to verify the assumption. The results showed that miR-18a was bound to the 3' UTR of Kdm5b (Figure 4B). Moreover, we observed that both the mRNA and protein levels of Kdm5b were significantly increased by miR-18a suppression but statistically decreased by miR-18a overexpression (P<0.01) (Figures 4C-4E). All the data demonstrated that Kdm5b was a target gene of miR-18a and was negatively regulated by miR-18a.
(A) Kdm5b was predicted as a potential target gene of miR-18a; (B), luciferase reporter assay confirmed that miR-18a was bound to the 3' untranslated region (UTR) of Kdm5b; (C), Real-time PCR and Western blot (D and E) were performed to verify the regulation relationship between miR-18a and Kdm5b.
miR-18a may mediate H3K4me3 expression
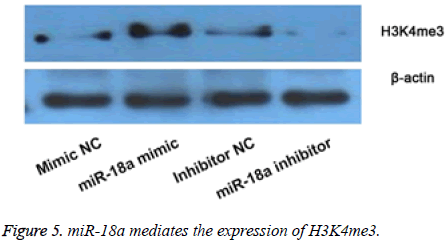
Then we observed the effects of miR-18a aberrant expression on level of H3K4me3 by WB (Figure 5). The results showed that compared with their corresponding groups, miR-18a mimic obviously increased the level of H3K4me3, and while the miR-18a inhibitor markedly inhibited the expression of H3K4me3. The results implied that the effects of miR-18a on the inner development might be by regulating the expression of H3K4me3.
The expression of H3K4me3 was detected by Western blot after overexpression or suppression of miR-18a. The results showed that overexpression of miR-18a increased the expression of H3K4me3, and while suppression of miR-18a showed opposite results.
Discussion
In the present study, we observed that the expression of miR-18a and Kdm5b was dynamically regulated in the developmental process of inner ear. During inner ear development, miR-18a was widely expressed in the cochlear hair cells, spiral ganglion, and vestibular sensory cells, and while the expression of Kdm5b was gradually decreased during inner ear development. In addition, the results confirmed that Kdm5b was a target gene of miR-18a. Moreover, the data demonstrated that miR-18a positively controlled the H3K4me3 level. Our results indicated that miR-18a may play significant roles in the development of inner ear by directly regulating the expression of Kdm5b and indirectly controlling the expression of H3K4me3.
The development and maturity of vertebrate inner ear requires a highly ordered and complex architecture, containing a multitude of different types of cell [2,21]. The developmental patterning mechanisms of the inner ear are varied and complex, involving numerous cellular factors and many signaling pathways [22]. It has been well acknowledged that miRNAs substantially contribute to the development and function of the organs [23]. Currently, an increasing number of studies have confirmed that some specific miRNAs might influence formation of sensory epithelia from the primitive otic neuroepithelium, as well as the development and function of inner ear hair cells [2,24]. In the present study, we focused on the functional role of miR-18a in the development of inner ear. miR-18a was reported in the development and morphogenesis of zebrafish inner ear [7]. Consistent with the previous study [7], we also confirmed that miR-18a may play an important role in the process of inner ear development in mice. Our data demonstrated that miR-18a was abundantly expressed in the inner ear spiral ganglion and Corti's organ at different development stages of inner ear. Similarly, we found that miR-18a was abundantly expressed in the hair cells.
Interestingly, our study meanwhile verified that Kdm5b was gradually decreased during the process of inner ear development. Kdm5b is a member of the JmjC domaincontaining histone demethylases, playing an important role in the process of DNA double-strand break (DSB) response in human cells [14,25]. Previous studies showed that Kdm5b is of great importance for normal embryonic development [26,27]. In addition, studies have suggested that Kdm5b is essential or dispensable for embryonic stem (ES) cell self-renewal and differentiation [28,29]. In line with the previous studies, our study also found that Kdm5b was responsible for the embryonic development. Considering the expression of miR-18a in the inner ear development, we assumed that there might be underlying regulation relationships between miR-18a and Kdm5b. As expected, Kdm5b was proved as a target gene of miR-18a and was also negatively regulated by miR-18a either in mRNA level or in protein level.
Additionally, it has been well known that Kdm5b catalyzes the removal of methyl groups from tri-, di-, and mono-methylated lysine 4 of histone H3 (H3K4me3/2/1) [30]. More recently, a study confirmed that Kdm5b plays critical roles in ES cell selfrenewal and differentiation by co-localizing with H3K4me3 near promoters and enhancers of active genes in ES cells [17]. It has been reported that H3K4me2 expression in the nuclei of hair cells, its decreasing for demethylation can leading to hairs cells’ loss [31]. In addition, a report showed the histone modifications H3K4me3 was subsequent silenced during the perinatal period, which reflected the therapeutic targets in hair cell regeneration [32]. In view of the relationship between Kdm5b and miR-18a/H3K4me3, therefore, we hypothesized that miR-18a could also regulate the expression of H3K4me3 in hair cells. As expected, our results confirmed that miR-18a positively regulated the expression of H3K4me3.
However, there are limitations in our study. Our results didn’t provide all the results of dynamic development process of every part in the inner ear. Hence, further studies with respect to the effects of aberrant expression of miR-18a and/or Kdm5b on the differentiation and development of inner ear and hair cells should be performed to confirm the results. In conclusion, our study suggests that the expression of miR-18a and Kdm5b was dynamically regulated in the developmental process of inner ear. miR-18a may play significant roles in the development of inner ear by directly regulating the expression of Kdm5b and by indirectly controlling the expression of H3K4me3.
Acknowledgement
The authors would like to acknowledge the research funding’s received under the grant numbers: 81271076 and 81200748.
References
- Li Y. Polymorphisms of heat shock protein 70 genes (HSPA1A, HSPA1B and HSPA1L) and susceptibility of noise-induced hearing loss in a Chinese population: A case-control study. PLoS One 2017; 12: p. e0171722.
- Doetzlhofer A, Avraham KB. Insights into inner ear-specific gene regulation: Epigenetics and non-coding RNAs in inner ear development and regeneration. Semin Cell Dev Biol 2016.
- Ni W. Wnt activation followed by Notch inhibition promotes mitotic hair cell regeneration in the postnatal mouse cochlea. Oncotarget 2016; 7: 66754-66768.
- Hailey DW. Fluorescent aminoglycosides reveal intracellular trafficking routes in mechanosensory hair cells. J Clin Invest 2017; 127: 472-486.
- Kwon DN. Oxidative stress and ROS metabolism via down-regulation of sirtuin 3 expression in Cmah-null mice affect hearing loss. Aging (Albany NY) 2015; 7: 579-594.
- Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development 2005; 132: 4653-4662.
- Friedman LM. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc Natl Acad Sci U S A 2009; 106: 7915-7920.
- Soukup GA. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev Biol 2009; 328: 328-341.
- Sacheli R. Expression patterns of miR-96, miR-182 and miR-183 in the development inner ear. Gene Expr Patterns 2009; 9: 364-370.
- Wang XR. MicroRNA expression in the embryonic mouse inner ear. Neuroreport 2010; 21: 611-617.
- Hirajima S. Clinical impact of circulating miR-18a in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer 2013; 108: 1822-1829.
- Luo Z. miR-18a promotes malignant progression by impairing microRNA biogenesis in nasopharyngeal carcinoma. Carcinogenesis 2013; 34: 415-425.
- Kee HJ. miR-18a-5p MicroRNA increases vascular smooth muscle cell differentiation by downregulating syndecan. Korean Circ J 2014; 44: 255-263.
- Li X. Histone demethylase KDM5B is a key regulator of genome stability. Proc Natl Acad Sci U S A 2014; 111: 7096-7101.
- Wang H. Transcriptional regulation of JARID1B/KDM5B histone demethylase by ikaros, histone deacetylase 1 (HDAC1), and casein kinase 2 (CK2) in B-cell acute lymphoblastic leukemia. J Biol Chem 2016; 291: 4004-4018.
- Rojas A. Epigenetic control of the bone-master runx2 gene during osteoblast-lineage commitment by the histone demethylase JARID1B/KDM5B. J Biol Chem 2015; 290: 28329-28342.
- Kidder BL, Hu G, Zhao K. KDM5B focuses H3K4 methylation near promoters and enhancers during embryonic stem cell self-renewal and differentiation. Genome Biol 2014; 15: R32.
- Li J. Cell-specific detection of miR-375 downregulation for predicting the prognosis of esophageal squamous cell carcinoma by miRNA in situ hybridization. PLoS One 2013; 8: e53582.
- Ye B. Cytosolic carboxypeptidase CCP6 is required for megakaryopoiesis by modulating Mad2 polyglutamylation. J Exp Med 2014; 211: 2439-2454.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402-408.
- Alsina B, Whitfield TT. Sculpting the labyrinth: Morphogenesis of the developing inner ear. Semin Cell Dev Biol 2016.
- Laos M. Indispensable role of Mdm2/p53 interaction during the embryonic and postnatal inner ear development. Sci Rep 2017; 7: 42216.
- Rebustini IT. A functional microrna screening method for organ morphogenesis. Curr Protoc Cell Biol 2017; 74: 19191-19197.
- Lewis MA. Exploring regulatory networks of miR-96 in the developing inner ear. Sci Rep 2016; 6: 23363.
- Han M. Histone demethylase lysine demethylase 5B in development and cancer. Oncotarget 2017; 8: 8980-8991.
- Albert M. The histone demethylase Jarid1b ensures faithful mouse development by protecting developmental genes from aberrant H3K4me3. PLoS Genet 2013; 9: e1003461.
- Catchpole S. PLU-1/JARID1B/KDM5B is required for embryonic survival and contributes to cell proliferation in the mammary gland and in ER+ breast cancer cells. Int J Oncol 2011; 38: 1267-1277.
- Xie L. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J 2011; 30: 1473-1484.
- Schmitz SU. Jarid1b targets genes regulating development and is involved in neural differentiation. EMBO J 2011; 30: 4586-4600.
- Klein BJ. The histone-H3K4-specific demethylase KDM5B binds to its substrate and product through distinct PHD fingers. Cell Rep 2014; 6: 325-335.
- He Y. Inhibition of H3K4me2 demethylation protects auditory hair cells from neomycin-induced apoptosis. Mol Neurobiol 2015; 52: 196-205.
- Stojanova ZP, Kwan T, Segil N. Epigenetic regulation of Atoh1 guides hair cell development in the mammalian cochlea. Development 2015; 142: 3529-3536.