Research Article - Biomedical Research (2018) Volume 29, Issue 12
Quality variations of extractable RNA in frozen samples of oral cancer (21 y); optimization of the extraction protocol
Cintia Chamorro M1,2, Mario Pérez-Sayans1,2*, Elena Padín Iruegas2,3, Xabier Marichalar Mendía4, José Suárez Peñaranda M2,5 and Abel García García1,2
1Department of Oral Medicine, Faculty of Medicine and Dentistry, Oral Surgery and Implantology Unit, Universidade de Santiago de Compostela, Spain
2Instituto de Investigación Sanitaria de Santiago (IDIS), Santiago de Compostela, Spain
3Department of Functional Biology, Vigo University, Spain
4Department of Nursery, Faculty of Medicine and Nursery, UPV/EHU, Spain
5Servicio de Anatomia Patológica, Hospital Clinico Universitario de Santiago, Choupana s/n Santiago de Compostela 15706, Spain
- *Corresponding Author:
- Mario Pérez-Sayans
Department of Oral Medicine
Faculty of Medicine and Dentistry
Oral Surgery and Implantology Unit
Universidade de Santiago de Compostela, Spain
Accepted date: March 09, 2018
DOI: 10.4066/biomedicalresearch.29-17-4031
Visit for more related articles at Biomedical ResearchAbstract
Access to sufficient tissue samples to develop a RNA study is complicated and in most cases a biobank is needed to preserve samples. Due to this it is necessary to assess how frozen tissue samples are influenced by the time they have been kept in storage. We have extracted RNA from 19 samples of oral squamous cell carcinoma (frozen from 1994 to 2015) and 31 samples of oral mucosa from healthy patients (extracted in 2015). The miRVana miRNA Isolation Kit (Ambion) was used to extract RNA. The extraction was performed following the manufacturer's instructions, except for the first part, sample disruption, for which we used a modified process. We determined the concentration of all samples, using NanoDrop 2000; while the RNA Integrity Number (RIN) was determined by NanoChip 6000 and the Agilent 2100 Bioanalyzer. The distribution of RIN and concentration is the same among location categories (p=0.696 and p=0.774, respectively). Results revealed a mean of RNA concentration lower in samples conserved for longer periods of time (p=0.006) but the RIN did not change with time storage (p=0.142). Our method was efficient to disrupt the tissue and to extract total RNA with good RIN and enough concentration. Although the concentration obtained from frozen tissue samples decreased with the passage of time, the RIN did not change.
Keywords
Total RNA, Biobank, Squamous cell carcinoma, Frozen tissue
Introduction
The techniques developed to increase our knowledge on the human genome have evolved rapidly over the past years, making them more accessible for research studies. The molecular tools used to explore tissue expression profile, such as microarrays, qRT-PCR (quantitative reverse transcription polymerase chain reaction) or ultrasequencing platforms, which allow global and specific evaluation of the genome and epigenome, are critical for the development of customized therapies for patients, especially in the case of cancer patients. These tools generally require the collection of fresh tissue as a source of high-quality RNA [1].
The fresh non-frozen tissue is the preferred specimen of current studies, since it produces a large quantity of highquality nucleic acids and proteins that paraffin-embedded tissue cannot achieve [2]. RNA is a very labile biomolecule and is susceptible to degradation by exogenous and endogenous nuclease and nonspecific degradation (heat, pH elevation and extensive periods of tissue storage) [3]. In general, the high-quality of total RNA is assessed on the basis of purity and integrity. RNA integrity is calculated by analyzing subunits 28S and 18S of the ribosomal RNA by means of electrophoresis chips, while the purity is determined by calculating the absorption ratio (A260 nm/A280 nm and A260 nm/A230 nm) using spectrophotometry. Most authors agree that the best strategy to evaluate RNA integrity is the estimation of RIN (RNA Integrity Number) using Agilent Bioanalyzer chips [4].
The quality of the studies on differential expression with RNA depends on the amount of material used and how robust the extraction method is in the reproduction of results [5]. All the platforms require a minimal concentration of RNA for their analysis, which usually ranges between 10-200 ng/microliter. Certain platforms, such as ultrasequencing, also recommend that the quality of the total RNA has a minimum RIN of 6 [1].
There are several different technologies used to isolate RNA from a frozen sample, however, there is an increasingly growing trend to use extraction kits that come at a high cost and may require additional equipment, depending on the chosen method. Knowing which factors influence the frozen samples is important so we do not lose time and money on materials that could be useless. Furthermore, the analysis of each of the samples in isolation as they are collected would come at a very high cost for the study, since these kits are available for a specific amount of samples and have an expiration date [6]. For the aforementioned reasons, public biobanks represent a very valuable tool thanks to their efforts to store and preserve patient samples.
However, one of the obstacles we have faced when extracting RNA from frozen tissue from the oral cavity is the processing of the sample. Most of the protocols recommend freezing in liquid hydrogen after harvesting the sample and within the least amount of time possible. In our study, the samples had been simply dried frozen or in O.C.T. compound for many years, no other media like liquid nitrogen or Invitrogen RNA later had been used. Therefore, these samples were perfect to discern if the passage of time affects RNA quality. Extracting total RNA from oral tissue also resulted in the need to seek an alternative to the first part of the sample processing, namely the disruption, so the protocol was modified accordingly to use a hand tool and a diamond drill.
The purpose of our study was to evaluate the quality of total RNA of the frozen oral cancer samples, measuring RIN and concentration, and optimizing the extraction protocol. For this purpose, we have divided the samples into two groups: oral cancer (1994-2015) and oral mucosa healthy control samples (2015). This division was made considering the specific conditions surrounding the cells of squamous cell carcinomas [7], to thus determine if the characteristics of the tissue have an influence on the RIN or the concentration of extractable total RNA.
Material and Methods
Samples
Nineteen oral cancer samples were obtained from patients who underwent surgery at Maxillofacial Surgery Unit of Complejo Hospitalario Universitario de Santiago de Compostela (Santiago Teaching Hospital) from January 1994 to December 2015. Thirty one oral mucosa samples were collected from healthy patients that visited the same service for the extraction of their third molar in 2015. Written informed consent was obtained from all patients prior to sampling. Patients with oral cancer were diagnosed by two independent pathologists, as oral squamous cell carcinoma, and the localization of samples were oral mucosa, soft palate, retromolar area, and the tongue and floor mouth. All the samples were stored at -80°C in the Santiago de Compostela Biobank (SERGAS). OCT© (Optimal Cutting Temperature) freezing cuts were developed on each sample, 14 cuts in 14 micrometers. The present study was approved by the Clinical Research Ethics Committee of Galicia Ref. 2015/132 (CEIC).
Extraction protocol
The miRVana miRNA Isolation (Ambion) kit was used for the extraction of total RNA. The protocol recommends immediate freezing upon the collection of the tissue sample in liquid nitrogen and maintaining the sample frozen at -80°C. To begin the extraction, the kit recommends dissolving the sample in a mortar, until its complete pulverization and adding a lysis buffer. However, when using this method, we encountered a series of difficulties that compromised the process: a) The sample fell out of the mortar easily; b) The sample thawed before total pulverization; c) A great amount of time was needed to achieve manual pulverization of the sample; d) It was difficult to obtain complete manual pulverization of the sample; e) It was impossible to use samples that had been frozen without using liquid nitrogen. The last issue (e) especially affected our study, since none of the samples preserved by the Biobank had been frozen in liquid nitrogen.
An alternative that the kit contemplated was the use of a motorized homogenizer for soft tissues. Due to the aforementioned problems, we used one following the manufacturer’s recommendations, but we still did not obtain complete homogenization; in this case, we believe it was due to the fibrous nature of the tissue used in this study (oral cancer samples).
Seeking alternatives for tissue pulverization, we decided to use a hand tool (Dremel 200 Series) with 1 mm-diameter diamond drills with a long 45 mm handle (2 mm Dremel diamond drill) and modified the initial step of the protocol by adding 300 μl of lysis buffer, which was included in the miRVana miRNA Isolation (Ambion) kit, freezing them once more in the same tube in which the sample had been provided by the Biobank of the Santiago Teaching Hospital.
Once the buffer froze completely, we used the diamond drill and the hand tool to obtain complete pulverization. The fact that we froze the lysis buffer with the sample prevented the latter from turning within the liquid medium without becoming disrupted. With this change we obtained the optimal state of the sample to continue with the procedure, following the manufacturer’s recommendations. Once the extraction was completed, the concentration and the purity of each sample was measured using NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA); and the RIN using Agilent RNA 6000 NanoChips in the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA).
All methods were performed in accordance with the relevant guidelines and regulations.
Statistical analysis
For the univariate description we used basic descriptive statistics: mean, standard deviation, frequency and percentage. To compare the means of extracted total RNA and RIN among the two sample groups, cases and controls, we used the U Mann Whitney test. For the analysis of the relationship between total RNA and RIN regarding the time during which the sample was preserved, we used non-parametric statistics through the Spearman correlation. For the analysis of the relationship between total RNA concentration and RIN regarding the type of tissue, we used non-parametric statistics by means of the Kruskal Wallis test. The significance level used was 5%, considering the value of p ≤ 0.05 indicative of a statistically significant relationship.
Results
Descriptive analysis
Distribution of age, gender, tumor stage and year of storage in oral cancer group are reported in Table 1. Distribution of age and gender and other variables in control group are reported in Table 2. The mean age of the 19 cases was 56.78 y, with a range of 31-75 y. Of the 19 cases, 6 were women (31.6%) and 13 were men (68.4%). The mean age of the 31 controls was 41.06 y, with a range of 18-85 y. Of the 31 controls, 12 were women (38.7%) and 19 were men (61.3%). 19 cases were primary intraoral tumors, 9 on the tongue (47.3%), 6 on the oral mucosa (31.5%), 1 in the retromolar area (5%), 2 in the floor mouth (10.5%), 1 in the soft palate (5%). In the oral cancer group 6 tissue samples (31.58%) had been frozen for less than 5 y (after 2010), whereas 13 samples had been frozen for over 5 y (68.42%).
| Case | Year storage | Localization | Stage | Gender | Age | Concentration (ng/µL) | RIN |
|---|---|---|---|---|---|---|---|
| A | 1997 | Oral mucosa | IV | Female | 69 | 191 | 2.9 |
| B | 1999 | Tongue | I | Male | 46 | 40 | 3.3 |
| C | 1999 | Soft palate | IV | Male | 57 | 43 | 3.6 |
| D | 2000 | Tongue | IV | Male | 47 | 61 | 1.1 |
| E | 1999 | Floor mouth | IV | Male | 38 | 48.57 | 6.9 |
| F | 2006 | Tongue | IV | Male | 70 | 300.86 | 6.5 |
| G | 2001 | Tongue | II | Male | 31 | 210.87 | 7.1 |
| H | 2014 | Tongue | IV | Female | 68 | 281.5 | 7 |
| I | 2014 | Retromolar area | IV | Male | 58 | 653.5 | 8.4 |
| J | 2014 | Oral mucosa | II | Male | 65 | 2149.53 | 8.9 |
| K | 2012 | Tongue | II | Male | 68 | 2655.2 | 8.5 |
| L | 1994 | Tongue | II | Male | 70 | 10 | 6.5 |
| M | 1997 | Oral mucosa | IV | Male | 39 | 374 | 8.1 |
| N | 1998 | Oral mucosa | IV | Male | 61 | 322 | 8.8 |
| O | 1998 | Oral mucosa | IV | Female | 41 | 36 | 6.8 |
| P | 1999 | Tongue | IV | Female | 67 | 229 | 10 |
| Q | 2001 | Tongue | IV | Male | 58 | 478 | 8.8 |
| R | 2015 | Tongue | II | Female | 75 | 528 | 8.3 |
| S | 2015 | Floor mouth | IV | Female | 51 | 972.27 | 8 |
Table 1. Distribution of age, gender, tumor stage and year of storage, concentration and RIN in oral cancer group.
| Control | Age | Gender | Concentration (ng/µL) | RIN |
|---|---|---|---|---|
| 1 | 24 | Female | 64.27 | 8.6 |
| 2 | 29 | Female | 444 | 8.2 |
| 3 | 24 | Female | 344.5 | 6,6 |
| 4 | 31 | Female | 220 | 6.5 |
| 5 | 20 | Female | 92.38 | 8,3 |
| 6 | 32 | Female | 195.54 | 8.9 |
| 7 | 33 | Male | 403.23 | 7 |
| 8 | 58 | Male | 163.34 | 7 |
| 9 | 49 | Male | 263.55 | 9.6 |
| 10 | 26 | Male | 208.52 | 9 |
| 11 | 18 | Male | 266.17 | 8.6 |
| 12 | 85 | Female | 596 | 8.8 |
| 13 | 53 | Female | 430 | 10 |
| 14 | 40 | Male | 632 | 8.8 |
| 15 | 40 | Male | 433 | 9,6 |
| 16 | 18 | Female | 202 | 8,6 |
| 17 | 80 | Female | 349 | 10 |
| 18 | 56 | Male | 1525 | 8.8 |
| 19 | 22 | Male | 347 | 7,3 |
| 20 | 21 | Male | 291 | 9.3 |
| 21 | 36 | Female | 212 | 9.2 |
| 22 | 18 | Female | 274 | 10 |
| 23 | 38 | Male | 96.04 | 9.8 |
| 24 | 56 | Male | 2930.6 | 8.1 |
| 25 | 52 | Male | 269.22 | 8.4 |
| 26 | 39 | Male | 111.07 | 8.9 |
| 27 | 74 | Male | 335.1 | 9,7 |
| 28 | 33 | Male | 300.04 | 9.7 |
| 29 | 18 | Male | 82.41 | 8.7 |
| 30 | 75 | Male | 1706 | 9,6 |
| 31 | 75 | Male | 1552 | 8.2 |
Table 2. Distribution of age, gender, concentration and RIN in control samples.
The results of RIN and concentration of total RNA in both groups are reported in Tables 1 and 2. The analysis of the 19 cases revealed a mean RIN of 6.81 (± 2.4035) and a mean concentration of 504.43 (± 717.757) ng/microliter. The mean RIN of the 31 controls was 8.70 (± 0.9941) and the concentration was 494.81 (± 615.073) ng/microliter. The Mann Whitney U test was used to compare the means of the cases and the controls, revealing a value of p=0.49 for RNA concentration and p=0.001 for RIN. The RIN is significantly superior in the control samples, in comparison to the oral cancer cases.
Association between tumor location and RNA quality
The RIN mean, the concentration mean and the standard deviation results for each location of the case samples can be observed in Table 3.
| Location | Number of samples | Average RIN (standard deviation) | Concentration mean in ng/µL (standard deviation) |
|---|---|---|---|
| Tongue | 9 | 7.4 (2.6739) | 237.69 (783.92079) |
| Oral mucosa | 6 | 7.3 (2.661) | 954.62 (984.34) |
| Floor of mouth | 2 | 7.4 (0.7778) | 510.42 (653.15453) |
| Soft palate | 1 | 3.6 | 43 |
| Retromolar area | 1 | 8.4 | 653.5 |
Table 3. Mean RIN and concentration by location.
When comparing the RIN averages by location, we obtained a significance level of 0.696 (Kruskal-Wallis test), therefore the distribution of RIN is the same among location categories.
When comparing the concentration averages by location, we obtained a significance level of 0.774, so we did not find a relation with extracted total RNA from the samples.
Association between RIN and the year in which the samples were frozen
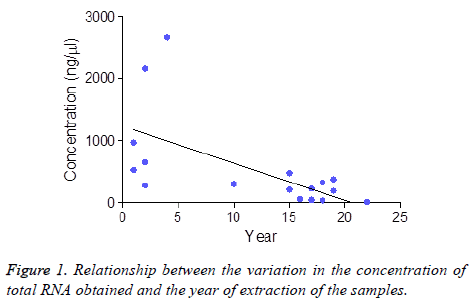
The Spearman correlation links the influence of the passage of time in the cases with total RNA concentration of -0.632 (p=0.006) (Figure 1) and a RIN of -0.350 (p=0.142). When dichotomizing the “freezing period” variable in samples of less and more than 5 y, in regards to concentration, there still are statistically significant differences (p=0.001 mean concentration in samples under 5 y: 1206.67 ng/microliter and over 5 y 180.33 ng/microliter). However, when comparing this dichotomized variable (5-y freezing period) with the RIN, we did not find statistically significant differences (p=0.087).
Discussion
The information on transcriptomes provides a fundamental vision of biological processes. The extraction of quality RNA is a complicated step and the preservation and extraction protocols have to be adjusted in many cases [4].
The automated methods for the extraction of RNA in fresh non-frozen tissue of the oral mucosa have shown better results than manual methods [8]. However, despite this, there are several factors that we must consider and that condition the studies: a) In many cases we must use frozen tissue; b) Not all laboratories have automated extraction technology; and, lastly, c) The acquisition of this material comes at a cost. All these factors have led us to find alternatives to make RNA extraction accessible both methodologically as financially.
The modification we developed, namely using the hand tool (Dremel 200 Series) and the diamond drill (2 mm Dremel diamond drill) implied an improvement in terms of time, since the sample can be totally homogenized in two minutes; but also financially, since drills can be sterilized and can be purchased at an affordable price. Additionally, the results obtained with this change reaffirm the appropriateness of its use.
In this study, we have proven that the concentration of extractable total RNA of frozen samples decreased with the passage of time (p=0.006). We haven't found articles mentioning the concentration of extractable total RNA related to the passage of time, but we have found studies documenting RNA degradation. Certain studies revealed RNA fragmentation after 5 y, despite having preservation conditions of -700°C or -800°C [9-11].
In our study, when dichotomizing the “freezing period “variable, dividing them between those stored for less than and more than 5 y, in terms of concentration we still found statistically significant differences (p=0.001 mean concentration in samples under 5 y: 1206.67 ng/microliter and over 5 y 180.33 ng/microliter). However, when comparing this dichotomized variable (5-y freezing period) in terms of RIN, we did not find statistically significant differences (p=0.087). These results require a greater sample size, although the Bioanlayzer technology does not indicate a minimum concentration for Nano Chips, it does recommend a concentration between 50-500 ng/microliter, and some of our samples showed a concentration lower than 50 ng/microliter.
Walter et al. extracted RNA from thyroid tumors preserved at different times, up to 10 years, and found no changes in RIN due to the passage of time, as was the case of Rudloff et al., who analyzed pancreatic tissue that had been frozen for 7 y (from 2001 to 2008) [12,13].The degradation is probably most influenced by the handling of the sample prior to freezing, rather than the amount of time during which it had been frozen.
In the study by Stiekema et al., part of the esophagus tissue was frozen in liquid nitrogen after the extraction and another part of the extracted tissue was frozen two hours after surgery. The results of the RIN showed an average of 6.7 for the tissue that was frozen immediately and 2.5 for the tissue that was frozen two hours later [14]. Other studies point to the ischemic or the specific characteristics of the tissue, which would affect the quality of the sample since, for example, the samples of the gastrointestinal tract in general seem to have lower quality RNA when compared to other organs. Some authors explain this fact with the presence of digestives or bacterial colonization [15].
The mirVanamiRNA Isolation kit was chosen as an isolation method because it allows analyzing all the RNAs of a sample, including those containing fewer than 200 nucleotides and, although there are other methods allowing the isolation of the RNA, for historical reasons mirVanamiRNA Isolation is the most extended kit [5].
Traditionally, the amount of RNA that can be extracted from tissue changes depending on the type of tissue, however, we haven’t found statistically significant differences when relating the concentration or the RIN of the oral cancer samples with the type of tissue they came from [16]. In fact, in contrast with what one would expect, the oral mucosa and the retromolar area were samples from which we obtained a higher concentration of total RNA in oral mucosa and retromolar area (oral mucosa=954.62 ng/microliter; retromolar=653.50 ng/ microliter) compared to the tongue (mucosa=237.69 ng/ microliter). An interesting result on which we have found no evidence up until now, is that the RIN extracted from the samples of healthy controls was higher than average compared to what was extracted from the samples of the oral cancer (p=0.001). This confirmed our initial suspicions on the influence of the circumstances that wrap the cancer tissue and RNA integrity.
The techniques of the transcriptome analyses require high RNA concentration and quality. For many authors, the paraffin-stored samples were a poor source of RNA, since the extracted nucleotic acids were fragmented and chemically modified. In our study, some samples had a RIN that would not be suitable for sensitive downstream analysis protocols such us qPCR, however, recent studies suggest that, for certain processes, lack of a minimum RIN is not incompatible with obtaining results in differential expression studies using ultrasequencing technology [17]. Other studies recommend that the analytic paraffin-based methods should only be considered as an alternative if there is no fresh non- frozen tissue [6].
Conclusions
The use of a hand tool and a diamond drill has proven to be a totally efficient RNA extraction method, with which we were able to obtain adequate RNA concentration and RIN. Extractable total RNA of the samples of frozen tissue decreases with the increase of preservation time. The RIN of the extracted total RNA of the frozen tissue samples was not influenced by the sample preservation time. The extractable RIN of the samples was significantly higher in healthy controls when compared to oral cancer samples. The fact that the tissue used was from different oral locations did not influence the results in terms of concentration and RIN of total RNA.
References
- Micke P. Biobanking of fresh frozen tissue: RNA is stable in nonfixed surgical specimens. Lab Invest 2006; 86: 202-211.
- Kalmar A. Gene expression analysis of normal and colorectal cancer tissue samples from fresh frozen and matched formalin-fixed, paraffin-embedded (FFPE) specimens after manual and automated RNA isolation. Methods 2013; 59: 16-19.
- Muyal JP, Muyal V, Kaistha BP, Seifart C, Fehrenbach H. Systematic comparison of RNA extraction techniques from frozen and fresh lung tissues: checkpoint towards gene expression studies. Diagn Pathol 2009; 4: 1596-1594.
- Perez-Portela R, Riesgo A. Optimizing preservation protocols to extract high-quality RNA from different tissues of echinoderms for next-generation sequencing. Mol Ecol Resour 2013; 13: 884-889.
- Mraz M, Malinova K, Mayer J, Pospisilova S. MicroRNA isolation and stability in stored RNA samples. Biochem Biophys Res Commun 2009; 390: 1-4.
- Ma R. Superficial scrapings from breast tumors is a source for biobanking and research purposes. Lab Invest 2014; 94: 796-805.
- Eckert AW. Clinical relevance of the tumor microenvironment and immune escape of oral squamous cell carcinoma. J Transl Med 2016; 14: 826-828.
- Van der Vorst S. Automated cell disruption is a reliable and effective method of isolating RNA from fresh snap-frozen normal and malignant oral mucosa samples. Clin Chem Lab Med 2009; 47: 294-301.
- von Teichman A, Storz M, Dettwiler S, Moch H, Schraml P. Whole genome and transcriptome amplification: practicable tools for sustainable tissue biobanking? Virchows Arch 2012; 461: 571-580.
- Shabihkhani M. The procurement, storage, and quality assurance of frozen blood and tissue biospecimens in pathology, biorepository, and biobank settings. Clin Biochem 2014; 47: 258-266.
- Lou JJ. A review of room temperature storage of biospecimen tissue and nucleic acids for anatomic pathology laboratories and biorepositories. Clin Biochem 2014; 47: 267-273.
- Walter MA. Extraction of high-integrity RNA suitable for microarray gene expression analysis from long-term stored human thyroid tissues. Pathology 2006; 38: 249-253.
- Rudloff U. Biobanking of human pancreas cancer tissue: impact of ex-vivo procurement times on RNA quality. Ann Surg Oncol 2010; 17: 2229-2236.
- Stiekema J, Cats A, Boot H, Langers AM, Balague Ponz O, van Velthuysen ML. Biobanking of fresh-frozen endoscopic biopsy specimens from esophageal adenocarcinoma. Dis Esophagus 2015; 29: 1100-1106.
- Zeugner S, Mayr T, Zietz C, Aust DE, Baretton GB. RNA quality in fresh-frozen gastrointestinal tumor specimens-experiences from the tumor and healthy tissue bank TU Dresden. Recent Results Cancer Res 2015; 199: 85-93.
- Tissot C. Analysis of miRNA content in total RNA preparations using the Agilent 2100 bioanalyzer. Agilent Technologies, Palo Alto, Calif, USA 2008.
- Hedegaard J. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS One 2014; 9: 98187.