Research Article - Journal of Clinical Pathology and Laboratory Medicine (2018) Journal of Clinical Pathology and Laboratory Medicine (Special Issue 1-2018)
Production of protease enzyme from fish guts using Pseudomonas fluorescens, Enterobacter cloacae and Bacillus megaterium.
Fajingbesi AO1, Abbas Abel Anzaku2*, Akande M1, Ikwue Adakole Emmanuel3, Ombugadu Akwashiki4
1Department of Biological Sciences, Fountain University Osogbo, Osun State, Nigeria
2Department of Microbiology, Federal University Lafia, Nasarawa State, Nigeria
3Department of Microbiology, School of Midwifery Makurdi, Benue State, Nigeria
4Department of Zoology, Federal University Lafia, Nasarawa State, Nigeria
- Corresponding Author:
- Abbas Abel Anzaku
Department of Microbiology
Fesderal University
Lafia Nsarawa state
Nigeria
Tel: +234 34 243 744
E-mail: humbleabel2016@yahoo.com
Accepted Date: January 29th, 2018
Citation: Fajingbesi AO, Anzaku AA, Akande M, et al. Production of protease enzyme from fish guts using Pseudomonas fluorescens, Enterobacter cloacae and Bacillus megaterium. J Clin Path Lab Med. 2018;2(1):1-7.
Abstract
Food industry wastes are an essential environmental contamination source. A large number of microorganisms including bacteria, yeast and fungi produce different types of enzymes like protease, lipase, chitinase, chitosan and ligninolytic enzyme having high biotechnological interest such as food processing, detergent, textile, pharmaceutical products, and medical therapy. Microbial synthesis of enzymes has been reported to be influenced by various factors such as carbon sources, nitrogen sources, and operating parameters like the temperature and pH. This research has been done in order to develop methods to convert these wastes into useful products. Three proteolytic organisms (Pseudomonas fluorescens, Enterobacter cloacae and Bacillus megaterium) were used in the production of protease and the most effective production was observed at a temperature of 45°C and pH 9. P. fluorescens produced protease best at 3rd production of 48 and 72 hours, E. cloacae produced protease best at 3rd production at 72 hours and B. megaterium produced best at 24 hours of 3rd production. P. fluorescens, E. cloacae and B. megaterium can produce protease and can therefore be used in the degradation of fish waste. These enzymes can also be used medically to control blood clotting target for degrading pathogenic proteins.
Keywords
Protease enzyme, Fish guts, Pseudomonas fluorescens, Enterobacter cloacae, Bacillus megaterium.
Introduction
Aquaculture has grown significantly in a number of European Union countries over recent years, and it is essentially an economic development within small and medium-sized businesses in remote areas where alternative employment is rather limited. This has been particularly obvious in marine aquaculture, Atlantic salmon in Scotland, Norway and Ireland, sea bass and sea bream (Sparusaurata) in the Mediterranean and mussel (Mytilusedulis) farming by line or raft in Ireland, Spain and France. This overall turn was improved because of a general decrease in catchable wild fish stocks and a rise in public demand for finfish and shellfish resources [1].
While capture fisheries fall short of world demand, annual consumption of seafood has been increasing, multiplying in three decades. Obviously, just as man no longer depends on hunting, he can no longer depend mainly on fishing. As of today, aquaculture provides over a quarter of the world’s seafood supply, a figure the FAO expects will improve by 50% by the year 2030 [2]. Global production of fish and shrimp has been increasing steadily over the last decade and this trend is expected to continue. Of the estimated 131 million tonnes of fish produced in 2000 in the world, nearly 74%, 97 million tonnes was used for direct human consumption. The remainder about 26% was utilized for series of non-food products, majorly for reduction to meal and oil. As a highly perishable good, fish has a significant requirement for processing. In 2000, more than 60% of total world fisheries production underwent some form of processing [3-10].
Food industry wastes are an essential environmental contamination source. Research has been done in order to develop methods to convert these wastes into useful products [11-20]. Possibly, more than 50% of the residue from the total fish capture is not used as food and involves about 32 million tonne's of waste (Figures 1-3).
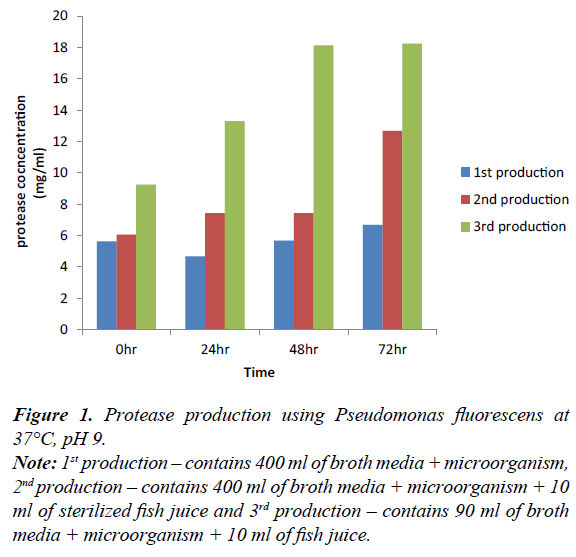
Figure 1: Protease production using Pseudomonas fluorescens at 37°C, pH 9.
Note: 1st production – contains 400 ml of broth media + microorganism, 2nd production – contains 400 ml of broth media + microorganism + 10 ml of sterilized fish juice and 3rd production – contains 90 ml of broth media + microorganism + 10 ml of fish juice.
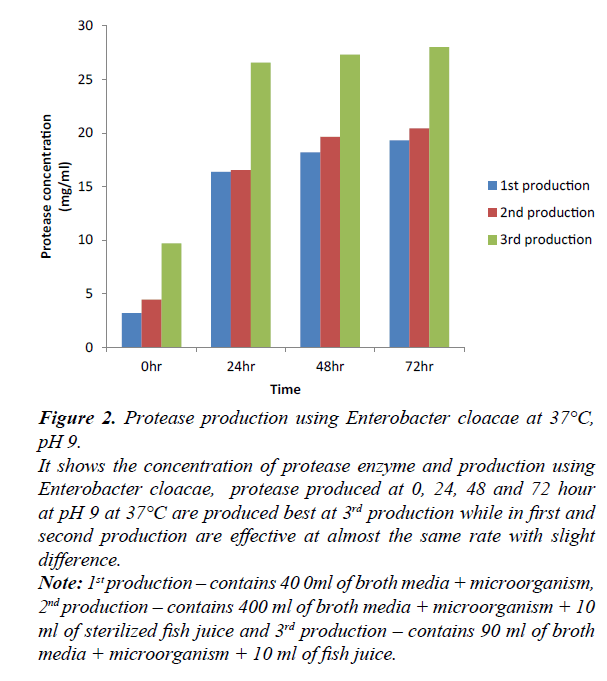
Figure 2: Protease production using Enterobacter cloacae at 37°C, pH 9.
It shows the concentration of protease enzyme and production using Enterobacter cloacae, protease produced at 0, 24, 48 and 72 hour at pH 9 at 37°C are produced best at 3rd production while in first and second production are effective at almost the same rate with slight difference.
Note: 1st production – contains 40 0ml of broth media + microorganism, 2nd production – contains 400 ml of broth media + microorganism + 10 ml of sterilized fish juice and 3rd production – contains 90 ml of broth media + microorganism + 10 ml of fish juice.
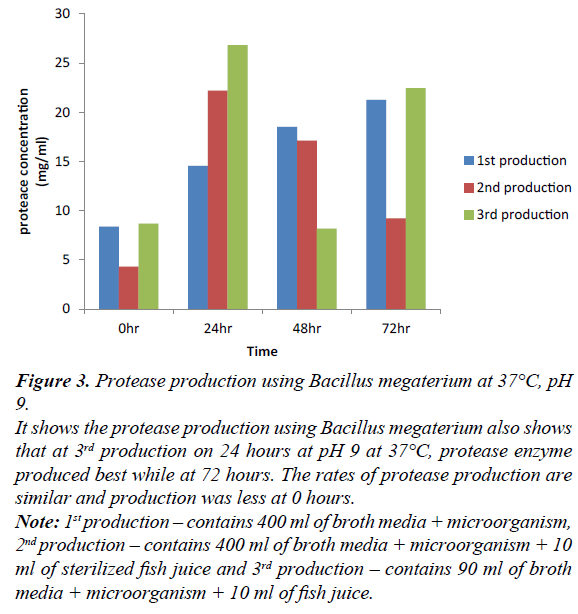
Figure 3: Protease production using Bacillus megaterium at 37°C, pH 9.
It shows the protease production using Bacillus megaterium also shows that at 3rd production on 24 hours at pH 9 at 37°C, protease enzyme produced best while at 72 hours. The rates of protease production are similar and production was less at 0 hours.
Note: 1st production – contains 400 ml of broth media + microorganism, 2nd production – contains 400 ml of broth media + microorganism + 10 ml of sterilized fish juice and 3rd production – contains 90 ml of broth media + microorganism + 10 ml of fish juice.
Figure 1 shows the concentration of protease enzyme produced using Pseudomonas fluorescens, at pH 9 at 72 hours in 3rd production; the microorganism tends to produce the highest concentration of enzyme, followed by 2nd production at 0 hr, 24 hr, and 48 hr at pH 9 and at 37°C respectively. First production can also increase with time at same temperature and pH also.
Interestingly, fish by-products provide an excellent source for microbial growth, which can be exploited in producing various metabolites like the lysine, enzymes, etc. [21-25]. A large number of microorganisms including bacteria, yeast and fungi produce different groups of enzymes like protease, lipase, chitinolytic and ligninolytic having high biotechnological interest such as food processing, detergent, textile, pharmaceutical products, medical therapy etc. [26-30]. Microbial synthesis of enzymes has been reported to be influenced by various factors such as carbon sources, nitrogen sources, and operating parameters like the temperature and pH [31-35]. Generally, agro-industrial residues mainly composed of complex polysaccharides are considered the best substrates for microbial growth and enzyme production [36-40]. The economies of this process are governed mostly by the cost and availability of a suitable carbon and nitrogen sources. The problem of waste generation and management in most cities especially in developing has become one of the intractable environmental problems facing urban centres. This situation could be attributed to low level of technology that is not sophisticated enough to handle the high rate of waste generation [41-45]. Human population and ruralurban migration has increased through urbanization, natural increase rate and industrialization, yet the service rendered is not sizeable enough to control the high level of solid waste generated in urban areas and these has contributed to a large extent, the menace and damaging effect of the urban environment [46-50]. Today, the rate at which waste is being generated is about 70% as compared to the total rate of its disposal which is 30%. No doubt [51-55] argued that waste could be anything which may not be directly useful or needed by man. In Nigeria, the dehumanizing effects of these circumstances in our urban lives and brighted environments has often been cited and noted as contributing causes of Nigerian urban decay [56-60]. Cross River State and Calabar in particular is not left out in this scenario despite the huge revenue allocated to waste disposal and management, the rate of waste generation and disposal is really alarming [61-65].
Therefore, the investigation of low cost substrates easily available such as fish processing wastes became a necessity opening new avenues for their effective production and utilization. In this study, the potential of different fish processing by-products tested as growth substrates for microbial enzyme production is presented. The aimed at producing protease enzyme using microorganism that is capable of degrading fish waste.
Materials and Methods
Collection of fish waste sample
Fish waste samples were collected in fresh conditions from Fountain University cafeteria. The waste material was sorted out and kept in big plastic bags and transported to the laboratory. The fish waste consists of the skin, scales, shell, fin, head, tail, frame and offal (guts, kidney and liver). The collected samples were taken to the back of the laboratory and were placed on the soil and were left to degrade.
Isolation and identification of bacteria from the soil
The soil was serially diluted by mixing 1 g in 9 ml distilled water, that gives 10-1 dilution. To the first tube of serially arranged 9 ml distilled water, 1 ml of 10-1 dilution was transferred which gives 10-2 dilution. Similarly, dilution was made up to 10-6. Total Bacteria Count (TBC) and Total Proteolytic Bacterial Count (TPBC) of each sample were determined using standard methods on Plate Count Agar, and Skim Milk Agar respectively. 0.1 ml of sample dilution 10-4, 10-5 and 10-6 was employed for plating. Isolates which shows clear zones on Skim Milk Agar were subjected to Gram staining. The isolates were further characterized based on their reactivity to various biochemical tests [66-70].
Selection of proteolytic isolate
Clear zones produced by the isolates on Skim Milk agar plates were measured. Thus, the proteolytic isolate which exhibit the highest proteolytic activity (determined by the extent of clear zone around the colony) was selected for further analysis [71-75].
Effect of pH on selected proteolytic isolate
Skim milk agar plates with different pH (7, 9, 13) were prepared and inoculated with the cultured isolate for determining the effect of pH on the proteolytic activity of the isolate [76].
Effect of temperature on selected proteolytic isolate at pH 9
Skim Milk Agar plates with pH 9 were prepared and inoculated with the selected isolate and incubated at different temperatures (25°C, 35°C and 45°C) for determining the effect of temperature on its proteolytic activity.
Protease production
The selected proteolytic isolates and the isolates were grown separately in production media containing 2% NH2SO4 (w/v), 1% K2HPO4(w/v), 1% KH2PO4(w/v), 0.4% MgSO4.7H2O(w/v), 0.01% MnSO4(w/v), 0.01% Fe2SO4.7H2O(w/v), 1% yeast extract(w/v), 10% peptone(w/v) with pH 9 maintained at 35°C for 24 to 72 hours in a shaking incubator(140 rpm). The second production media to be subjected for protease production was the sterilized remaining part of homogenized fish juice. The third production media to be subjected for protease production was 90 ml production media vortexes mixing thoroughly with 10 ml sterilized homogenized fish juice [77].
Extraction of enzyme and dialysis
The cells were harvested by centrifugation at 10000 rpm for 10 minutes at 4°C. The precipitate containing the cell debris was discarded and supernatant treated with 70% Ammonium sulphate and allowed to stand at 4°C overnight. Then the precipitate was centrifuged at 10000 rpm for 20 mins at 4°C. The resulting precipitate was dissolved in 0.9% NaCl and labeled as crude protease extract. The same process was used for the remaining above-mentioned media too. The crude enzyme was partially purified using ammonium sulphate (60% saturation). The precipitated enzyme was used as a crude protease.
Proteolytic activity profile
The proteolytic activity of crude protease extract was assayed at pH4-10 using the following systems-Citrate phosphate buffer (pH 4 and 5), phosphate buffer (pH 6-8), Glycine-NaOH buffer (pH 9 and 10). The assay mixture will consist of 1.25 ml of respective buffers, 0.5 ml of casein as substrate and 0.25 ml of enzyme extract. The reaction was stopped by adding 3 ml of 5% TCA after holding it at 37°C for 30 minutes. The final mixture was filtered and TCA soluble peptides in filtrate were quantified by measuring the absorbance at 540 nm using Biurets working solution’s method.
Results
Soil samples were serially diluted and dilutions of 10-4, 10-5, and 10-6 were plated on Nutrient Agar, and Skim Milk Agar in which the total bacteria count, total proteolytic bacteria count, and their morphological characteristics were presented in Tables 1-3.
| Dilution Plate | Colony Count (CFU/ml) |
|---|---|
| 10-4 | 1.05 × 105 |
| 10-5 | 6.1 × 104 |
| 10-6 | 2.2 × 106 |
Table 1. Total bacterial count on nutrient agar plate.
| Dilution Plate | Colony count (CFU/ml) |
|---|---|
| 10-4 | 0.8 × 104 |
| 10-5 | 0.5 × 105 |
| 10-6 | 0.2 × 106 |
Table. 2 Total proteolytic bacterial count on skimmed milk agar.
| Morphology | A | B | C |
|---|---|---|---|
| Colour | Creamy | Creamy | Creamy |
| Elevation | Raised | Flat | Flat |
| Surface | Moist | Dry | Moist |
| Edges | Irregular | Clear | Round |
Table 3. Morphological characteristics on skimmed milk agar.
Microorganism that can utilize the milk present in the skimmed milk agar were subjected to various biochemical tests as shown in Table 4 the isolates were suspected to be Pseudomonas fluorescens, Enterobacter cloacae and Bacillus megaterium. Table 5 shows the clear zone measurement of each organism. At 24-hour P. fluoresens shows that there was a little clear zone compared to 48 and 72 hours respectively. A gradual increase in the clear zone was also observed at 24 hours up to 72 hours in E. cloacae. B. megaterium at 24 hour shows no clear zone while at 48-72 hour show clear zones. These may be due to late lag phase of the microorganism (Tables 6 and 7).
| Biochemical test | A | B | C |
|---|---|---|---|
| Gram’s reaction | -ve rod | -ve rod | +ve rod |
| Catalase | +ve | +ve | +ve |
| Oxidase | -ve | -ve | -ve |
| urease | +ve | -ve | -ve |
| Nitrate reduction | -ve | Nil | -ve |
| Indole | +ve | +ve | -ve |
| Starch hydrolysis | +ve | +ve | +ve |
| actose | -ve | +ve | -ve |
| citrate | +ve | +ve | -ve |
| fructose | +ve | Nil | +ve |
| Sucrose | Nil | Nil | +ve |
| xylose | +ve | nil | Nil |
| glucose | +ve | +ve | -ve |
| mannitol | +ve | nil | +ve |
| Methyl red | Nil | +ve | +ve |
| Hydrogen sulphide | nil | -ve | -ve |
| Voges proskauer | nil | -ve | +ve |
| Suspected organism | Pseudomonas fluorescens | Enterobacter cloacae | Bacillus megaterium |
Table 4. Tests carried out on isolates which shows clear zone on skimmed milk agar.
| Organisms | Clear zone at 24 hours | Clear zone at 48 hours | Clear zone at 78 hours |
|---|---|---|---|
| Radius (cm) | Radius (cm) | Radius (cm) | |
| P. fluoresens | 0.1 | 0.2 | 1.1 |
| E. cloacae | 0.3 | 4 | All cleared |
| B. megaterium | Nil | 4 | All cleared |
Table 5. Clear zones measurement of each isolates.
| Isolates | pH 7 | pH 9 | pH 13 |
|---|---|---|---|
| P. fluoresens | 0.6 | 3.5 | 1.5 |
| B. megaterium | 0.3 | 2.3 | 0.6 |
Table 6. Effect of ph on all the isolates.
| Isolates | Temp. 25°C | Temp. 35°C | Temp. 45°C |
|---|---|---|---|
| P. fluoresens | 0.6 | 0.4 | 0.9 |
| E. cloacae | 2 | 2.8 | 3.1 |
| B. megaterium | 2.3 | 3.4 | 3.2 |
Table 7. Effect of temperature on the isolates with sma of pH 9.
It shows the effect of pH the proteolytic isolate and each isolate were subjected to different pH of (7, 9 and 13) the isolate that shows the most effective was at pH 9 and has the widest clear zone and these indicates the level of alkalinity suitable for the growth of the microorganisms.
Discussion
Based on the outcome of this research, organisms capable of producing protease enzymes were identified according to the Tables 3 and 4 after the sample collection and degradation. They were isolated using nutrient agar and skimmed milk agar respectively. The growth on skimmed milk agar shows that microorganisms that can utilize protein are present. Biochemical test was carried out on them and microorganism suspected were Pseudomonas fluorescens, Enterobacter cloacae and Bacillus megaterium. The organisms gotten from this study is in agreement with the study. The effect of temperature and pH on the skimmed milk agar shows that pH 9 and 45°C were effective. Protease enzymes were produced at 0, 24, 48 and 72 hours and at three productions. The first production of protease enzymes using Pseudomonas fluorescens, at 72 hours during the 3rd production is best in all production.
Production of protease using Pseudomonas fluoresence, at 0, 24, 48 and 72 hours. 0 hour of 3rd production gradual increase in the level of protease concentration in the production. At 0 hour the 1st and 2nd production varies with increase in the 2nd production this is due to the difference in their constituent present in the media. 2nd and 3rd production varies due to the volume ratio which is lower in 3rd production.
Production of protease using Enterobacter cloacae, at 0, 24, 48 and 72 hours. At 0 hr, first and second production varies as a result of the difference in their constituent, second and third production varies as a result of difference in their volume ratio. At 24 hr, first and second productions were observed to be at the same level despite similar ratio and different constituent. Third and second productions were observed with increase in their result due to low volume ratio and similar constituent. At 24 hr, first and second production were observed to vary slightly with increase in second production, second and third production varies greatly due to difference in their volume ratio and similar constituent, first and third production varies greatly due to difference in constituent and volume ratio. At 72 hr, first and second production varies due to different constituent but similar volume ratio, second and third production varies due to same constituent with different volume ratio. The third production produces best at 0, 24, 48, and 72 hours with time.
Production of protease enzyme using Bacillus megaterium at 0, 24, 48, 72 hours. At 24 hr of third production shows the best protease production in all the production. At 0 hr, first and third production have equal rate compare to second production, this is due to different microorganism, constituent and volume ratio. At 24 hr, there was gradual increase in production. At 48 hr, first production shows more level of concentration compare to second and third production. At 72 hr, first and third production vary slightly, this may be due to different constituent and volume ratio compare to 24 hr. 24 hr of all the production using Bacillus megaterium produced best in second production. Comparing all the bacteria’s involved in protease production of this study; third production is suitable for the production of each microorganism.
Conclusion
In conclusion Pseudomonas fluorescens, Enterobacter cloacae and Bacillus megaterium are protease enzymes and can be isolated from the soil and can be using in both in the degradation of fish waste and also in the production of protease enzyme that can be useful in food industries, medically, pharmaceutical industries and even agriculturally. Fish waste should not be dumped anyhow because they are have created more good than harm, including the scales are beneficial to the world.
Recommendation
Fish waste should be recycled so as to produce enzyme that are beneficial to the world rather than disposing inappropriately and causing environmental pollution to the community. Utilization fish wastes, fish waste as silage, fish meals for livestock agriculture, pharmaceuticals etc. are also the beneficial use of fish waste. The protease enzyme can be used in industries in the production of baby food like Nan, laundry detergents etc. Protease enzymes are good medically by controlling blood clotting target for degrading pathogenic proteins.
References
- Ackman RG, Mcleod C. Total lipids and nutritionally important fatty acids of some Nova Scotia fish and shelfish products. CanadianInstitute Food Science Technology. 1988;24:390-98.
- Aidos I, A-van-der P, Boom RM, et al. Upgrading of maatjes herring byproducts: production of crude fish oil. J Agri Food Chem.2001;49:3697-704.
- An HK, Park BY, Kim DS. Crab shell for the removal of heavy metals from aqueous solutions. Water Resources. 2001;35:3551-556.
- Batista I. Recovery of proteins from fish waste products by alkaline extraction. Euro Food Res Technol. 1999;210:84-9.
- Baum B, Parker GH. Solid waste disposal, vol. 1 incinerations and landfill, Michigan Ann Arober Science Publishers incorporated. 1973.
- Beerli EL, Beerli KM, Logato PVR. Silagemacida de resíduos de truta (Oncorhyncusmykiss) com autilização de ácidomuriático. CiênciaAgrotecnológica. 2004;28:195-98.
- Ben-Rebah F, Frikha F, Kammoun W, et al. Culture of Staphylococcus xylosus in fish processing by-product-based media for lipase production. Lett Appl Microbiol. 2008;47:549-554.
- Bhunia B, Dutta D, Chaudhari S. Selection of suitable carbon, nitrogen and sulphate source for the production of alkaline protease by Bacillus licheniformis NCIM– 2042. Notulae Scientia Biologicae. 2010;2:56-59.
- Bimbo AP, Crowther JB. Fish meal and oil: current uses. J Am Oil Chem Sociol. 1992;69:221-227.
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Canadian JBiochem Physiol. 1959;37:911-917.
- Borderias JA, Mateos PM. Restructured fish products. Alimentaria.1996;269:53-62.
- Borghes R. Avaliaçãofísico-química e biológica das silagensácSida, biológica E enzimáticaelaboradas com descarte e resíduo do beneficiamento da tilápia do nilo (Oreochromisniloticus). Dissertação (Mestrado) – Escola Superior de Agricultura “Luiz de Queiroz”, Universidade de São Paulo, Piracicaba, Brasil. 2004;108.
- Burrows R, Chenoweth H. Evaluation of three types of rearing ponds. Research Report 39, U.S. Department of Interior, Fish and Wildlife Service, Washington, D.C. USA. 1970.
- Coello N, Montiel E, Concepcion M, et al. Optimization of a culture medium containing fish silage for L-lysine production by Corynebacterium glutamicum. Bioresource Technol. 2002;85:207-11.
- Danzo BJ. Environmental xenobiotics may disrupt normal endocrine function by interfering with the binding of physiological ligands to steroid receptors and binding proteins. Environmental Health Perspect. 1997;105:294-301.
- Edu N. Environmental waste and management: Ushie printers and publishing Co. Limited Calabar. 2003.
- Esakkiraj P, Austin JebaDhas G, Palavesam A, et al. Media preparation using tuna-processing wastes for improved lipase production by shrimp gut isolate Staphylococcus epidermidis CMST Pi2. Biochemistry Biotechnology. 2010;160:1254-1265.
- Fagbenro O, Jauncey K. Physical and nutritional properties of moist fermented fish silage pellets as a protein supplement for tilapia (Oreochromisniloticus). Animal Feed Sci Technol. 1998;71:11-18.
- Faid M, Zouiten A, Elmarrakchi A, et al. Biotransformation of fish waste into a stable feed ingredient.Food Chemistry.1997;60:13-18.
- Food and Agricultural Organization of the United Nations (2002) FAO Fisheries Statistical Yearbook. 2002;27-39.
- Food and Agricultural Organization of the United Nations La situation mondiale des pêcheset de l’aquaculture (2006). Département des pêcheset de l’aquaculture, Rome. 2007.
- Food and Agricultural Organization of the United Nations. Animal feed resources information system. J Sci Food Agri.2003;25:369-79.
- Fernandes TF, Miller KL, Read PA. Monitoring and regulation of marine aquaculture in Europe. J Appl Ichthyol. 2000;16:138-43.
- Ferraz L. Aproveitamento do resíduo do beneficiamento da tilápia do nilo (Oreochromisniloticus) paraobtenção de silagem e óleocomosubprodutos. Dissertação (Mestrado)– Escola Superior De Agricultura “Luiz De Queiroz”, Universidade De São Paulo, Piracicaba, Brasil. 2004;78.
- Ferraz L, Borghesi R, Brum A. Ácidosgraxosemsilagem de resíduos do processamento da tilapia do nilo (Oreochromisniloticus). In: Congresso Latino Americano de Nutrição Animal - CLANA, 2. Anais. São Paulo:CBNA, CD-ROM. 2006.
- Gassara F, Brar SK, Tyagi RD, et al. Screening of agro-industrial wastes to produce ligninolytic enzymes by Phanerochaete chrysosporium. Biochem Engineer J. 2010;49:388-394.
- Gildberg A. Enzymes and bioactive peptides from fish waste related to fish silage, fish feed and fish sauce production. J Aqua Foods Product Technol. 2004;13:3-11.
- Gowen RJ. Aquaculture and the environment. In: Aquaculture and the Environment. DePauw N, Joyce J (eds) Ghent: European Aquaculture Society Special Publications. 1991;16:30-8.
- Hammoumi A, Faid M, El-Yachioui M, et al. Characterization of fermented fish waste used in feeding trials with broilers. Process Biochem. 1998;33:423-27.
- Hasan F, Shah AA, Hameed A. Industrial applications of microbial lipases. Enz Microbiol Technol. 2006;39:235-251.
- Heras H, Mcleod CA, Ackman RG. Atlantic dogfish silage vs. herring silage in diets for Atlantic salmon (Salmosalar): Growth and sensory evaluation of fillets. Aquaculture. 1994;125:93-103.
- Kacem M, Sellami M, Kammoun W, et al. Seasonal variations of chemical composition and fatty acid profiles of viscera of three marine species from the Tunisian coast. Aqua Food Prod Technol. 2011;20:233-46.
- Karim AA, Bhat R. Fish gelatin: Properties, challenges, and prospects as an alternative to mammalian gelatins. Food Hydrocolloids. 2009;23:563-76.
- Khan MAA, Hossain MA, Hara K, et al. Effect of enzymatic fish-scrap protein hydrolysate on gelforming ability and denaturation of lizard fish Sauridawaniesosurimi during frozen storage. Fisheries Sci.2003;69:1271-80.
- Kim JS, Shahidi F, Heu MS. Characteristics of salt-fermented sauces from shrimp processing by-products. J Agri Food Chem. 2003;51:784-92.
- Kim SK, Mendis E. Bioactive compounds from marine processing by-products-a review Food Res Int. 2006;39:383-93.
- Kirk TK, Farrell RL. Enzymatic “combustion”: The microbial degradation of lignin. Ann Rev Microbiol. 1987;41:465-501.
- Knorr D. Recovery and utilization of chitin and chitosan in food processing waste management. Food Technol.1991;45:114-122.
- Kompiang IP. Fish silage: Its prospect and future in Indonesia. Indonesia Agri Res Development J. 1981;3:9-12.
- Kristinsson HG, Rasco BA. Fish protein hydrolysates: production, biochemical, and functional properties. Crit Rev Food Sci Nutr. 2000;40:43-81.
- Kumaran E, Mahalakshmipriya A, Sentila R. Effect of fish waste based Bacillus Protease in silver recovery from waste X-Ray Films. Int J Curr Microbiol Appl Sci.2013;2:49-56.
- Larsen T, Thilsted SH, Kongsback K, et al. Whole small fish as a rich calcium source. Brit J Nutr. 2000;83:191-196.
- Laufenberg G, Kunz B, Nystroem M. Transformation of vegetable waste into value added products. Bioresource Technol. 2003;87:167-198.
- Midlen A, Redding T. Environmental Management for Aquaculture. London: Chapman & Hall.1998;223.
- Morimura S, Nagata H, Uemura Y, et al. Development of an effective process for utilization of collagen from livestock and fish waste Process Biochemistry. 2002;37:1403-12.
- Negroni G. Management optimization and sustainable technologies for the treatment and disposal/reuse of fish farm effluent with emphasis on constructed wetlands. World Aqua. 2000;31:16-19.
- Nogueira Junior S, Negri Neto A, Tsunechiro A, et al. Alimentação animal: realidade e perspectivas (Coleçãocadeias de produção da aqüicultura) São Paulo: Secretaria da Agricultura e Abastecimento. 1997;95.
- Nurullah A. Production of extracellular protease in submerged fermentation by Bacillus licheniformis ATCC 12759. Afr J Biotechnol. 2012;11:1729-35.
- http://www.oaerre.napier.ac.uk
- Oetterer M. Proteínas do pescadoprocessamento com intervençãoprotéica. In: Oetterer M, Regitanod´Arce, M.A.; Spoto MHF Fundamentos de Ciência e Tecnologia de Alimentos. Barueri: Manole. 2006;99-134.
- Ohba R, Deguchi T, Kishikawa M, et al. Physiological functions of enzymatic hydrolysates of collagen or keratin contained in livestock and fish waste. Food Sci Technol Resources. 2003;9:91-3.
- Palaniyappan M, Vijayagopal V, Renukal V. Screening of natural substrates and optimization of operating variables on the production of pectinase by submerged fermentation using Aspergillus niger MTCC 281. Africa Journal Biotechnology. 2009;8:682-686.
- Pandey A, Selvakumar P, Soccol CR. Solid state fermentation for the production of industrial enzymes. Curr Sci. 1999;77:149-162.
- Perea A, Ugalde U, Rodriguez I. Preparation and characterization of whey protein hydrolysates: Application in industrial whey bioconversion processes. Enz Microbial Technol. 1993 15:418-423.
- Pillay TVR. Aquaculture and the Environment. London: Blackwell Scientific Publications, UK. 1991;26-45.
- Potter D, Tatterson I, Wignall J. Preliminary studies of two technique for the removal of oil from fish silage using commercial equipment. In: Connell JJ. Adv Fish Sci Technol. Surrey: FAO Fishing News Books. 1979;338-43.
- Rabea WI, Badawy MET, Stevens CV. Chitosan as antimicrobial agent: Applications and mode of action. Biomacromolecules 2003;4:1457-1465.
- Rodehutscord M, Pfeffer E. Effects of supplemental microbial phytase on phosphorus digestibility and utilization in rainbow trout. Water Sci Technol. 1995;31:141-47.
- Rodríguez S, Toca JL. Industrial and biotechnological applications of laccases: a review. Biotechnol. 2006;24:500-513.
- Rubio-Rodriguez N, Beltran S, Jaime I. Production of omega-3 polyunsaturated fatty acid concentrates: A review. Inn Food Sci Emergency Technol.2010;11:1-12.
- Ruiz-Duenas FJ, Danzo BJ. Environmental xenobiotics may disrupt normal endocrine function by interfering with the binding of physiological ligands to steroid receptors and binding proteins. Environmental Health Perspect. 1997;105:294-301.
- Shahidi F, Arachchi JKV, Jeon Y. Food applications of chitin and chitosans. Trends Food Sci Technol. 1999;10:37-51.
- Sharma R, Chisti Y, Banerjee UC. Production, purification, characterization and applications of lipases.Biotechnol Advanced. 2001;19:627-662.
- Skorupa K, Sikorski ZE. Potential for utilization of low-grade fish material for production of microbiological peptones. Przem Spoz. 1993;47:159-61.
- Sule RO. Solid and faecal waste disposal urban Development planning framework the Nigeria experience;. thumbprint IntI Company. 2004.
- Summerfelt ST, Alder PR, Glenn DM, et al. 5th International Conference on Wetland Systems for Water Pollution Control, Vienna. 1996.
- Tahajod ASR, AG. Seafood waste potential to support antimicrobial compound production by lactic acid bacteria. IFT Annual meeting. 1996;31-2.
- Tidwell JH, Allen GL. Fish as food: Aquaculture’s contribution. Ecological and economic impacts and contributions of fish farming and capture fisheries. EMBO Reports. 2001;21:958-963.
- Triki-Ellouz Y, Ghorbel B, Souissi N. Biosynthesis of protease by Pseudomonas aeruginosa MN7 grown on fish substrate. Microbiol Biotechnol. 2003;19:41-45.
- Under kofler LA, Barton RR, Rennert SS. Microbial process report: production of microbial enzymes and their applications. Appl Microbiol. 1958;6:212-21.
- Vanquez JA, Gonzalez MP, Murado MA. Peptones from auto hydrolyzed fish viscera for nisin and pediocin production. J Biotechnol.2004;112:299-311.
- Vazquez JA, Docasal SF, Miron J, et al. Proteases production by two Vibrio species on residuals marine media. Industrial Biotechnol.2006;33:661-68.
- Vendruscolo F, Albuquerque PCM, Streit F, et al. Apple pace: A versatile substrate for biotechnological applications. Crit Rev Biotechnol. 2008;28:1-12.
- Waites MJ, Morgan NL, Rockey JS, et al. Industrial Microbiology an Introduction London: Blackwell Publisher. 2008.
- Wiefels RC. Panorama da aqüiculturamundial. Infopesca Internacional.1998;19-27.
- Yang JK, Tzeng IL, Wang SL. Production and purification of protease from a Bacillus subtilis that can deproteinize crustacean wastes. Enzyme Microbiol Technol. 2000;26:406-413.
- Yoshida H, Terashima M, Takahashi Y. Production of organic acids and amino acids from fish meat by sub-critical water hydrolysis. Biotechnology Programe. 1999;15:1090-94.