Research Article - Biomedical Research (2017) Volume 28, Issue 5
Probiotic therapy alleviates hyperuricemia in C57BL/6 mouse model
Tong Cao1, Xiaoyu Li1, Tao Mao1, Hua Liu1, Qingxi Zhao1, Xueli Ding1, Changgui Li2, Lijuan Zhang3, Zibin Tian1*1Department of Gastroenterology, the Affiliated Hospital of Qingdao University, Qingdao 266003, P.R. China
2Shandong Provincial Key Laboratory of Metabolic Disease, the Affiliated Hospital of Qingdao University, Qingdao 266003, P.R. China
3Institute of Cerebrovascular Diseases, the Affiliated Hospital of Qingdao University, Qingdao 266003, P.R. China
- *Corresponding Author:
- Zibin Tian
Department of Gastroenterology
The Affiliated Hospital of Qingdao University, P.R. China
Accepted date: October 26, 2016
Abstract
Aim: The present study is to investigate differences in gut bacteria especially bifidobacteria and lactobacilli between C57BL/6 mice with and without hyperuricemia, as well as the effect of probiotic therapy with bifidobacteria and lactobacilli on hyperuricemia-associated serum markers over time in the mouse model.
Methods: The mouse model of hyperuricemia was established by using C57BL/6 mice and a reported protocol. DNA was extracted from fecal samples collected from C57BL/6 mice. Specific primers for 16S rRNA gene were designed for PCR-based detection of bifidobacteria and lactobacilli, respectively. Uric acid (UA) level, lipopolysaccharides (LPS) level, and xanthine oxidase (XO) activity in serum were measured.
Results: The amount of bifidobacteria and lactobacilli in fecal samples from hyperuricemic mice was significantly decreased, while UA level, XO activity, and LPS level were significantly increased in hyperuricemic mice compared to those of normal mice. These changes were attenuated after five weeks of probiotic therapy.
Conclusion: The present study demonstrates the relationship between hyperuricemia and gut microbial changes over time and the effect of probiotic therapy on hyperuricemia in mouse model.
Keywords
Intestinal flora, Hyperuricemia, Uric acid, Lipopolysaccharide, Xanthine Oxidase.
Introduction
Uric acid (UA) is an end product of purine nucleoside metabolism in humans, which is regulated by balancing its production in the liver and urinary excretion [1-4]. Unlike other mammals, hominoids have high serum UA levels due to the lack of the urate-degrading enzyme uricase, which sometimes leads to hyperuricemia [5,6]. It has been suggested that serum UA should be kept below 7 mg/dL to prevent hyperuricemia, which is a clinically important risk factor for cardiovascular diseases, chronic kidney disease and gout [7,8]. Intestinal microorganisms play various roles in human health including metabolism of complex fiber and fats, as well as immune regulation [9]. Intestinal microbial imbalance increases lipopolysaccharide (LPS) levels in blood circulation, induces chronic inflammation [10], and is associated with metabolic syndrome (MS), obesity, and insulin resistance (IR) [11]. Numerous studies support the role of intestinal microbiome in regulating body weight and energy metabolism as well [12-16].
In healthy individuals, about 70% UA is excreted through kidneys and the remaining 30% is excreted or decomposed by intestinal flora [17,18]. As a metabolic disease, hyperuricemia is closely associated with increased production and reduced excretion of UA. However, the impact of reduced excretion of UA on the decomposition of UA by intestinal flora in hyperuricemia patients is unknown. Moreover, little work has been conducted to investigate the relationship between hyperuricemia and intestinal flora. Abnormal level of serum UA is a clinical marker for hyperuricemia. Xanthine oxidase (XO) is a key enzyme in the production of UA, and increased activity of XO can lead to increased UA production. LPS is a metabolite of intestinal flora. The abnormal level of LPS in blood circulation induces inflammation in many diseases accompanied by increased XO activity. Therefore, the pathological relationship between hyperuricemia and intestinal flora could be monitored by measuring UA level, LPS level, and XO activity in serum. In the present study, we employ high purine diet and administer intraperitoneal injection of potassium oxonate to induce consistent high levels of UA in C57BL/6 mice as hyperuricemia model [19,20]. We further use probiotics to modulate intestinal microbiome. The proportion of bifidobacteria and lactobacilli in intestinal flora is quantified, while LPS level, UA level and XO activity were measured to investigate the relationship between intestinal flora and hyperuricemia.
Materials and Methods
Animals
C57BL/6 mice (18-22 g) were purchased from the Animal Center of Shandong Gout Laboratory, Qingdao, China. All animals were handled according to the guidelines provided by the Animal Care and Use Committee at Shandong Province. Experimental protocols for animal use were reviewed and approved by the animal ethics committee of Qingdao University. Animals were housed at 26 ± 3°C with a 12 h light/12 h dark cycle.
Thirty mice were randomly divided into three groups (n=10 each) as follows: normal group (NG), hyperuricemia group (HG) and hyperuricemia with probiotic therapy group (HPG). NG was fed a normal diet (laboratory chow and water ad libitum) with saline (0.5 ml) by intragastric administration once a day, while the other two groups were fed high-purine diet (laboratory high-purine chow and water ad libitum) and administered intraperitoneal injection of potassium oxonate 300 mg/kg once a day, with saline (0.5 ml) for HP and probiotics (0.5 ml) for HPG (bifidobacteria and lactobacilli, 1.0 × 106 CFU each) by intragastric administration once a day.
DNA extraction
Fecal samples were collected into 1.5 ml tubes. To avoid contamination and microbial growth, fecal samples were put on ice immediately and stored at -80°C in less than an hour. Fecal samples were thawed and centrifuged at 12,000 rpm for 3 min. The pellet was collected for DNA extraction according to the manufacture’s instruction (QIAamp Fast DNA Stool Mini Kit; No. 148048456, Qiagen, Hilden, Germany). The quality of all DNA samples was analyzed by using 1% agarose gel electrophoresis. DNA concentration of each sample was quantified by taking 1.5 μL of the extracted DNA sample and measuring the OD using an ultraviolet spectrophotometry (Nanodrop ND2000, Thermo Scientific, Waltham, MA, USA). To ensure the repeatability and stability of PCR analysis, the concentration of all DNA samples were higher than 20 ng/μL according to the suggestions of a published report [21]. The OD values of all DNA samples were between 1.8-2.0, indicating the absence of RNA or protein cross contaminations.
Polymerase chain reaction (PCR)
Primers were designed based on 16S rRNA gene sequences available from GenBank using Primer Premier for Windows, version 5.0 (Premier Biosoft International, Palo Alto, CA) (Table 1). Primer specificities for their target organisms were checked with an online tool provided by the Ribosomal Database Project. Primers were synthesized commercially by Invitrogen Life Technologies (Thermo Fisher Scientific, Waltham, MA, USA). Each reaction mixture for PCR (50 μL) contained 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 1.5 mM MgCl2, 0.25 mM deoxynucleoside triphosphates, 0.5 M primer, 1 μL of bacterial template DNA, and 1 U of Taq DNA polymerase. Optimal annealing temperatures and specificity testing were done by using a Mastercycler gradient PCR instrument and a Techne Genius PCR instrument, respectively. PCR protocol consisted of 35 cycles with DNA denaturation at 95°C (1 min), followed by annealing (1 min) and elongation at 72°C (45 seconds); a final elongation step was performed at 72°C (5 min).
| Target organisms | Sequences (5’ to 3’) | Annealing temperature (°C) | References |
|---|---|---|---|
| Bifidobacteria | GGGTGGTAATGCCGGATG (forward) TAAGCCATGGACTTTCACACC (reverse) |
59 | 26 |
| Lactobacilli | CATCCAGTGCAAACCTAAGAG (forward) GATCCGCTTGCCTTCGCA (reverse) |
58 | 26 |
Table 1: PCR primers used in this study.
Quantitative real-time polymerase chain reaction (qRT-PCR)
According to references [22,23], iCycler iQ apparatus (Bio- Rad, Hercules, CA, USA) and iCycler Optical System Interface software (version 2.3; Bio-Rad, Hercules, CA, USA) was used for qRT-PCR. Each reaction was performed in duplicate in a volume of 20 μL in 96-well optical-grade PCR plates. Amplification reactions were done with iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) containing 3 mM MgCl2, 20 mM Tris HCl (pH 8.4), 50 mM KCl, deoxynucleoside triphosphate (200 μM), SYBR Green I, 10 nM fluorescein, 0.625 U iTaq DNA polymerase mixed with selected primer, and 1.6 μL of respective template DNA or water. Amplifications were done with the following protocol: one cycle at 95°C (3 min), 35 cycles of denaturation at 95°C (30 s), primer annealing (30 s), and one final cycle at 95 °C (30 s). Finally, melt curve analyses were made by slowly heating PCR mixtures from 55 to 95°C (1°C per cycle of 10 s) with simultaneous measurements of the SYBR Green I signal intensities. Quantification was done by using standard curves made from known concentrations of plasmid DNA containing respective amplicon for each set of primers.
Quantification of UA level, XO activity and LPS level in serum
Blood samples were collected from tail veins of mice on days 7, 14, 21, 28 and 35. Serum was obtained after centrifugation at 4,000 rpm for 10 min. Serum was used for measuring UA level, XO activity, and LPS level. UA was measured by a clinical automatic biochemical analyzer (AU5800, Beckman Coulter, Brea, CA, USA). Serum XO activity and LPS level were measured according to the manufacturer’s instructions (Xanthine Oxidase Activity Assay Kit; NO. MAK078, Sigma- Aldrich, St. Louis, MO, USA) and a previous publication [24], respectively.
Statistical analyses
Logarithms of fecal 16S rRNA gene copy numbers were used to achieve normal distribution, and means ± standard deviations were calculated. SPSS for Windows (version 10.0.7, IBM, Armonk, NY, USA) was employed for statistical analyses with one-way analysis of variance and Bonferroni multiple comparisons to compare the three groups with each other. P<0.05 was considered as statistical significance.
Results
The numbers of bifidobacteria and lactobacilli are reduced in hyperuricemic mice, and intragastric probiotic therapy increases the amount of bifidobacteria and lactobacilli in fecal samples. To determine rRNA gene copies, qRT-PCR was performed. The data showed that the number of rRNA gene copies of bifidobacteria from fecal samples in HG was significantly lower than that of NG on days 14, 21, 28 and 35, and that in HPG was significantly higher than that of NG and HG on all days (Table 2). Similarly, the number of rRNA gene copies of lactobacilli from fecal samples in HG was significantly lower than that of NG on days 14, 21, 28 and 35, and that in HPG was significantly higher than that of NG and HG on all days (Table 3). These results suggest that the numbers of bifidobacteria and lactobacilli are reduced in hyperuricemic mice, and intragastric probiotic therapy increases the amount of bifidobacteria and lactobacilli in fecal samples.
| Day 7 | Day 14 | Day 21 | Day 28 | Day 35 | |
|---|---|---|---|---|---|
| NG | 8.13 ± 0.20 | 8.14 ± 0.27 | 8.09 ± 0.32 | 8.10 ± 0.26 | 8.09 ± 0.47 |
| HG | 8.02 ± 0.33 | 7.66 ± 0.50# | 7.67 ± 0.33# | 7.71 ± 0.27# | 7.69 ± 0.25# |
| HPG | 8.57 ± 0.35#* | 8.76 ± 0.30#* | 8.82 ± 0.26#* | 8.98 ± 0.19#* | 8.94 ± 0.20#* |
Table 2: Quantification of bifidobacterium 16S rRNA genes in mouse fecal DNA preparations of normal group (NG), hyperuricemia group (HG) and probiotics-treated hyperuricemia group (HPG) at different time points (logCFU/g, means ± standard deviations, n=10).
| Day 7 | Day 14 | Day 21 | Day 28 | Day 35 | |
|---|---|---|---|---|---|
| NG | 7.12 ± 0.28 | 7.15 ± 0.30 | 7.19 ± 0.21 | 7.14 ± 0.23 | 7.10 ± 0.28 |
| HG | 7.09 ± 0.18 | 6.82 ± 0.23# | 6.78 ± 0.28# | 6.79 ± 0.24# | 6.80 ± 0.30# |
| HPG | 7.79 ± 0.20#* | 7.81 ± 0.25#* | 7.87 ± 0.19#* | 7.88 ± 0.17#* | 7.96 ± 0.16#* |
Table 3: Quantification of lactobacillium 16S rRNA genes in mouse fecal DNA preparations of normal group (NG), hyperuricemia group (HG) and probiotics-treated hyperuricemia group (HPG) at different time points (logCFU/g, means ± standard deviations, n=10).
Probiotic therapy reduces serum UA level in hyperuricemic mice
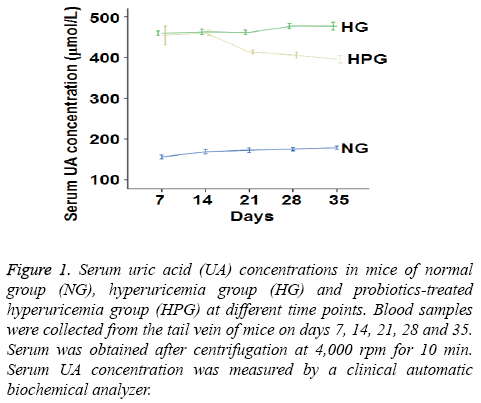
To measure serum UA level, a clinical automatic biochemical analyzer was used. The data showed that UA level was increased by at least 88% in HG mice compared to NG mice on days 7-35 (Figure 1). After intragastric probiotic therapy once a day, serum UA level in HPG mice was decreased by more than 18% compared to HG mice on days 21, 28 and 35 (Figure 1). However, serum UA levels in HPG mice were still significantly higher than that of NG. These results indicate that probiotics reduces serum UA level in hyperuricemic mice even though the UA level is still higher than that in normal mice.
Figure 1: Serum uric acid (UA) concentrations in mice of normal group (NG), hyperuricemia group (HG) and probiotics-treated hyperuricemia group (HPG) at different time points. Blood samples were collected from the tail vein of mice on days 7, 14, 21, 28 and 35. Serum was obtained after centrifugation at 4,000 rpm for 10 min. Serum UA concentration was measured by a clinical automatic biochemical analyzer.
Probiotic therapy decreases serum XO activity in hyperuricemic mice
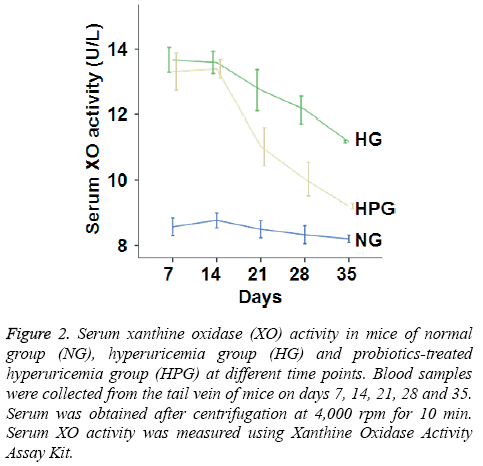
To determine XO activity, colorimetric assay was performed. The data showed that XO activity in HG mice was significantly increased compared to that in NG mice on days 7-35. Moreover, XO activity in HPG mice was significantly decreased compared to that in HG mice on days 21, 28 and 35. However, serum XO activity in HPG mice was still significantly higher than that in NG mice (Figure 2). The results suggest that probiotic therapy reduces the activity of XO, a key enzyme in the production of UA.
Figure 2: Serum xanthine oxidase (XO) activity in mice of normal group (NG), hyperuricemia group (HG) and probiotics-treated hyperuricemia group (HPG) at different time points. Blood samples were collected from the tail vein of mice on days 7, 14, 21, 28 and 35. Serum was obtained after centrifugation at 4,000 rpm for 10 min. Serum XO activity was measured using Xanthine Oxidase Activity Assay Kit.
Probiotic therapy decreases serum LPS level in hyperuricemic mice
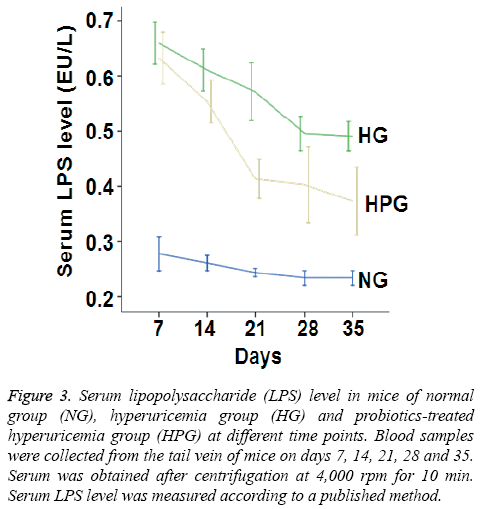
To investigate the metabolism of intestinal flora, serum LPS level was measured. The data showed that serum LPS level in HG mice was significantly increased compared to that in NG mice on days 7-35. By contrast, serum LPS level in HPG mice was significantly decreased compared to that in HG mice on days 14-35 (Figure 3). These results indicate that probiotic therapy decreases serum LPS level in hyperuricemic mice.
Figure 3: Serum lipopolysaccharide (LPS) level in mice of normal group (NG), hyperuricemia group (HG) and probiotics-treated hyperuricemia group (HPG) at different time points. Blood samples were collected from the tail vein of mice on days 7, 14, 21, 28 and 35. Serum was obtained after centrifugation at 4,000 rpm for 10 min. Serum LPS level was measured according to a published method.
Discussion
Hyperuricemia is a clinically important risk factor for cardiovascular diseases, chronic kidney disease and gout. At present, the clinical medicine used for hyperuricemia treatment has low drug tolerance, high drug dependence and multiple side effects due to lack of understanding of the complicated disease. Interestingly, imbalanced compositions of intestinal flora are associated with human metabolic diseases related to abnormal lipid and sugar metabolism. The amount of probiotics, especially bifidobacteria and lactobacilli, is decreased in these diseases [10-12,15,16]. As a metabolic disease, hyperuricemia is associated with increased production and reduced excretion of uric acid. However, the impact of reduced excretion of uric acid on decomposition of uric acid by intestinal flora of hyperuricemic patients is unknown. Therefore, we have hypothesized that hyperuricemia may also be linked to changes of probiotics in gut microbiome. To establish the mouse model of hyperuridemia, a high-purine diet and intraperitoneal injection of potassium oxonate at 300 mg/kg once a day were used to maintain a consistent high level of serum uric acid over 35 days. The data showed the reliability of the hyperuricemic mouse model.
Most of the studies on human gut bacteria use cultivation techniques to monitor bacterial populations, although it has been reported that less than 25% of the intestinal species are cultivable [24]. In the present study, we have used molecular biology techniques based on qRT-PCR analysis of 16S rRNA genes to determine the amount of probiotics, especially bifidobacteria and lactobacilli. The qRT-PCR technique has enabled the populations of bifidobacteria and lactobacilli to be directly quantified by using DNA isolated from fecal samples [25]. We observe that the numbers of rRNA gene copies of both bifidobacteria and lactobacilli in fecal samples are significantly lower in HG mice on days 14-35 compared to those in NG mice. The observation indicates that the populations of intestinal flora are changed in hyperuricemic mice, mainly in the reduction of the probiotics bifidobacteria and lactobacilli. Interestingly, probiotic therapy is widely used clinically in a variety of diseases associated with the reduction of probiotics such as bifidobacteria and lactobacilli. However, little work has been conducted on whether probiotics reduces serum uric acid levels. In the present study, intragastric probiotic therapy with both bifidobacteria and lactobacilli has restored the amount of the probiotics beyond the normal level. In addition, serum UA level in HPG is decreased by more than 18% compared to that in HG mice after day 21. Therefore, the application of probiotics reduces serum UA level in hyperuricemic mice.
Imbalance of intestinal flora is accompanied by an increase of LPS in blood circulation. High level of serum LPS induces chronic inflammation that is closely associated with many diseases such as metabolic syndrome, obesity, and insulin resistance [10,11]. In addition, serum LPS level is reduced when mice are treated with probiotics in a metabolic disorder model [26]. XO is a key enzyme in the production of uric acid. Chronic inflammation induced by increasing serum LPS level is often accompanied by increased XO activity [27]. In the present study, serum LPS level in HG mice is significantly increased compared to that in NG mice on days 7 to 35. Serum XO activity in HG mice is increased by more than 26% compared to that in NG mice on days 7 to 35. After day 14, serum XO activity and LPS level in HPG mice are decreased by almost 20% compared to those in HG mice after probiotic therapy. Therefore, these results show clearly that serum LPS level is increased in hyperuricemic mice and accompanied by increased serum UA level and XO activity in HG mice. Moreover, probiotic therapy decreases not only LPS level but also serum XO activity and UA level. Therefore, the present study provides novel insights in understanding the relationship between hyperuricemia and intestinal flora.
There are also some limitations in the present study. Firstly, probiotic therapy reduces serum UA level in hyperuricemic mice, but UA concentration is still significantly higher than that in NG mice. Moreover, whether serum UA level is related to the dosage of probiotics therapy is unclear. Secondly, the mouse model resembles high serum UA level in hyperuricemic human patients, but the cause of human hyperuricemia is much more complex than that of mouse model. Therefore, it is still unknown whether probiotics has therapeutic effect on human hyperuridemic patients. In conclusion, the present study demonstrates that hyperuricemia is associated with changed intestinal flora, mainly in the reduction of probiotics such as bifidobacteria and lactobacilli. Probiotic treatment increases the amount of bifidobacteria and lactobacilli in intestinal flora, and decreases UA level, LPS level, and XO activity in serum.
Acknowledgment
This research was supported by the Natural Science Foundation of China (Grant No. 91129706), and Taishan Scholar Fellowship of Shandong Province.
References
- Hopper DC, Scott GS, Zborek A, Mikheeva T, Kean RB, Koprowski H, Spitsin SV. Uric acid, a peroxynitrite scavenger, inhibits CNS inflammation, blood-CNS barrier permeability changes, and tissue damage in a mouse model of multiple sclerosis. FASEB J 2000; 14: 691-698.
- Masuo K, Kawaguchi H, Mikami H, Ogihara T, Tuck ML. Serum uric acid and plasma norepinephrine concentrations predict subsequent weight gain and blood pressure elevation. Hypertension 2003; 42: 474-480.
- Daimon M, Ji G, Saitoh T, Oizumi T, Tominaga M, Nakamura T, Ishii K, Matsuura T, Inageda K, Matsumine H, Kido T, Htay L, Kamatani N, Muramatsu M, Kato T. Large-scale search of SNPs for type 2 DM susceptibility genes in a Japanese population. BiochemBiophys Res Commun 2003; 302: 751-758.
- Tasic V, Janchevska A, Emini N, Sahpazova E, Gucev Z, Polenakovic M. Chronic kidney disease pediatric risk factors. Pril 2016; 37: 9-13.
- Keebaugh AC, Thomas JW. The evolutionary fate of the genes encoding the purine catabolic enzymes in hominoids, birds, and reptiles. MolBiolEvol 2010; 27:1359-1369.
- Dabbagh F, Ghoshoon MB, Hemmati S, Zamani M, Mohkam M, Ghasemi Y. Engineering Human Urate Oxidase: Towards Reactivating It as an Important Therapeutic Enzyme.Curr Pham Biotechnol 2015; 17: 141-146.
- Gliozzi M, Malara N, Muscoli S, Mollace V. The treatment of hyperuricemia. Int J Cardiol 2016; 213: 23-27.
- Schils R, Krzesinski JM. Hyperuricemia and potential risk of cardiovascular and renal diseases. Rev Med Liege 2016; 71: 262-268.
- Lai S, Zhou X. Inflammatory cells in tissues of gout patients and their correlations with comorbidities. Open Rheumatol Journal 2013; 7: 26-31.
- Kunitskaia NA, Kozina LS, Utkin AK, Utkina AA. The peculiarities of Chronic inflammation in elderly patients with gout and metabolic syndrome. AdvGerontol 2013; 26: 161-165.
- Roberfroid M, Gibson GR,Hoyles L, McCartney AL, Rastall R, Rowland I, Wolvers D, Watzl B, Szajewska H, Stahl B, Guarner F, Respondek F, Whelan K, Coxam V, Davicco MJ, Léotoing L, Wittrant Y, Delzenne NM, Cani PD, Neyrinck AM, Meheust A. Prebiotic effects: metabolic and health benefits. Br J Nutr 2010; 104: S1-S63.
- Vitetta L, Gobe G. Uremia and chronic kidney disease: the role of the gut microflora and therapies with pro-and prebiotics. MolNutr Food Res 2013; 57: 824-832.
- Ellekilde M, Krych L, Hansen CH, Hufeldt MR, Dahl K, Hansen LH, Sørensen SJ, Vogensen FK, Nielsen DS, Hansen AK. Characterization of the gut microbiota in leptin deficient obese mice-Correlation to inflammatory and diabetic parameters. Res Vet Sci 2014; 96: 241-250.
- Remely M, Aumueller E, Jahn D, Hippe B, Brath H, Haslberger AG. Microbiota and epigenetic regulation of inflammatory mediators in type 2 diabetes and obesity. Benef Microbes 2014; 5: 33-43.
- Joyce SA, Gahan CG. The gut microbiota and the metabolic health of the host. Cur OpinGastroenterol 2014; 30: 120-127.
- Zupancic ML, Cantarel BL, Liu Z, Drabek EF, Ryan KA, Cirimotich S, Jones C, Knight R, Walters WA, Knights D, Mongodin EF, Horenstein RB, Mitchell BD, Steinle N, Snitker S, Shuldiner AR, Fraser CM. Analysis of the gut microbiota in the old order Amish and its relation to the metabolic syndrome. Plos One 2012; 7: e43052.
- Hosomi A, Nakanishi T, Fujita T, Tamai I. Extra-renal elimination of uric acid via intestinal efflux transporter BCRP/ABCG2. Plos One 2012; 7: e30456.
- Krishnan E. Chronic kidney disease and the risk of incident gout among middle-Aged men: A seven-year prospective observational study. Arthritis Rheum 2013; 65: 3271-3278.
- Liu YW, Sun WF, Zhang XX, Li J, Zhang HH. Compound Tufuling Granules regulate glucose transporter 9 expression in kidney to influence serum uric acid level in hyperuricemia mice. Chin J Integr Med 2015; 21: 823-829.
- Hou PY, Mi C, He Y, Zhang J, Wang SQ, Yu F, Anderson S, Zhang YW, Wu XH. Pallidifloside D from Smilax riparia enhanced allopurinol effects in hyperuricemia mice. Fitoterapia 2015; 105: 43-48.
- Krishnan E. Chronic kidney disease and the risk of incident gout among middle-Aged men: A seven-year prospective observational study. Arthritis Rheum 2013; 65: 3271-3278.
- Bartosch S, Fite A, Macfarlane G. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderlypatients by using real-time PCR and effects of antibiotic treatment on the fecalmicrobiota. Appl Environ Microbiol 2004; 70: 3575-3581.
- Aparecida de Oliveira M. Quantification of Listeria mmocytogens in minimally processed leafy vegetables using a combined method based on enrichment and 16S rRNA real-time PCR. Food Microbiol 2010; 27: 19-23.
- Wang CR, Cheng YG, Zeng J. Effect of Intestinal Function-recovering Decoction on the bacterial and endotoxin translocation as well as MDA and SOD in serum of rats with multiple organ dysfunction syndrome Journal of Xi’an Jiaotong University 2009; 30: 119-123.
- Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, Doré J. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol 1999; 65: 4799-4807.
- Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, Burcelin R. Changes in gut microbiota control metabolic endotoxemia-induced inflammatiom in high-fat diet-induced obesity and diabetes in mice. Diabetes 2008; 57: 1470-1481.
- Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmée E, Cousin B, Sulpice T, Chamontin B, Ferrières J, Tanti JF, Gibson GR, Casteilla L, Delzenne NM, Alessi MC, Burcelin R. Metabolic endotoxemiaintiates obesity and insulin resistance. Diabetes 2007; 56: 1761-1772.