Case Report - Biomedical Research (2017) Volume 28, Issue 7
Prenatal diagnosis of small supernumerary marker chromosome 15: A case report
Bo Wang1#, Zhi Liu1#, Handong Fu2#, Haitao Chen3 and Jinou Xi1*1Clinical Laboratory, Hubei Maternal and Child Health Hospital, Wuhan, Hubei, China
2Central Laboratory, Xiaogan Hospital affiliated to Wuhan University of Science and Technology, Xiaogan, Hubei, China
3Department of Urology, Hubei Maternal and Child Health Hospital, Wuhan, Hubei, China
#These authors contributed equally to this work
- *Corresponding Author:
- Jinou Xi
Associate Chief Physician
Clinical Laboratory Hubei Maternal and Child Health Hospital
China
Accepted date: November 26, 2016
Abstract
Small Supernumerary Marker Chromosomes (sSMCs) are rare, being present in less than 0.08% of all pregnancies. The precise characterization of marker chromosomes is important for prenatal diagnosis and proper genetic counselling. Here we report a case of prenatally diagnosed de novo sSMC derived from chromosome 15. Our case emphasizes the importance of the modern aspects of array CGH in combination of thorough ultrasound examination to provide precise and rapid prenatal diagnosis.
Keywords
Karyotype analysis, Fluorescence in situ hybridization, Array comparative genomic hybridization, Chromosome 15, Prenatal diagnosis, Small supernumerary marker chromosomes
Introduction
Small Supernumerary Marker Chromosomes (sSMCs) are defined as structurally abnormal chromosomes that cannot be identified or characterized by conventional karyotype analysis and are generally equal in size or smaller than a chromosome 20 [1]. sSMCs are present in nearly 0.044% of live births and 0.075% of prenatal cases [2,3]. Approximately 77% of sSMCs arise de novo and 23% are inherited [2,3]. Nearly 70% of sSMCs are derived from the short arms and pericentromeric regions of acrocentric chromosomes [4]. Most (70%) of de novo sSMCs have no phenotypic effects [5]. However, in at least 30-50% of prenatally detected sSMC cases, the pregnancy is terminated [2,3], which means unnecessary abortions were induced in a certain percentage of potentially healthy children with sSMC. Therefore the precise characterization of marker chromosomes is crucial for prenatal diagnosis and proper genetic counselling. Here we present the molecular characterization of an sSMC derived from chromosome 15 in prenatal diagnosis.
Case Presentation
A 29-year-old primigravid underwent amnioinfusion at 20 weeks of gestation due to a Down’s syndrome risk of 1 in 90. Conventional cytogenetics revealed an abnormal karyotype, 47, XY, +mar with one sSMC detected in all metaphase, thereby making the possibility of mosaicism unlikely (Figure 1).
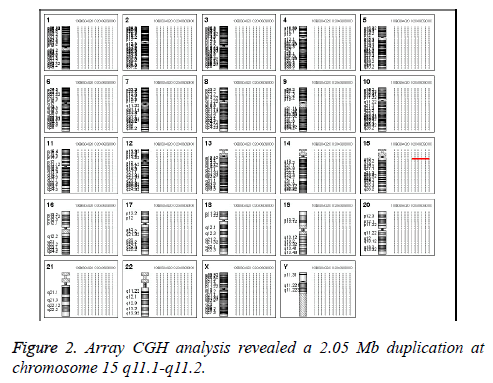
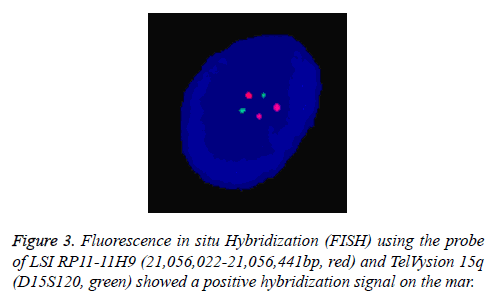
In Addition, the parental karyotypes were normal. In order to determine from which chromosome the sSMC comes, array Comparative Genomic Hybridization (aCGH) analysis was performed using a commercially available human wholegenome CGH array (Agilent Technologies, Santa Clara, CA, USA). DNA was extracted from peripheral blood as previously described [6,7]. A 2.05 Mb duplication was detected on the long arm of chromosome 15 (cytogenetic location 15q11.1- q11.2) (Figure 2). According to the Copy-Number Variation (CNV) database, this is a chromosomal polymorphic variation. The aCGH result was further confirmed by double-locus fluorescence in situ hybridization (FISH) (Figure 3) [8]. The FISH image demonstrated three signals for 15q11.1 (red) but two signals for 15q26.3 (green, control). To evaluate the foetal development, we performed a thorough ultrasound examination. It showed no facial dysmorphisms, structural abnormalities or Intrauterine Growth Restrictions (IUGRs). Based on these results, we suggested the couple to continue the pregnancy. At 39 weeks of gestation, a 3150 g female baby with a karyotype of 47, XY, +Mar was delivered. At one year of age, the baby was progressing normally.
Discussion
Identification of sSMC is one of the most difficult tasks in prenatal cytogenetics. Conventional karyotype analysis can detect numerical and structural chromosomal abnormalities but cannot determine the origin and genetic content of sSMCs. Traditional approaches for their characterization such as twenty-four color FISH, centromere or sub-centromere-specific multicolor FISH and micro-dissection followed by reverse FISH, are time-consuming and can result in ambiguous classification or misclassification of sSMC [9]. Recently, array CGH analysis has been shown to be highly accurate for rapid detection of chromosomal aneuploidies and sub-microscopic deletions or duplications on foetal DNA samples [10-13]. In this study, we validate that array CGH analysis provides an alternative to telomere FISH and disease-specific FISH in the cytogenetic diagnostic laboratory. Array CGH has the ability to detect DNA dosage imbalance including deletions and duplications in the euchromatic regions and is useful for the characterization of the origin and hereditary effects in the sSMC.
The correct characterization of gene deletions and duplications is a crucial point in order to identify the genotype phenotype correlation. In fact, entire and partial gene deletions/ duplications can produce a completely different phenotypic effect. The low copy repeats (LCRs) in chromosome 15q11- q13 have been recognized as Breakpoints (BP) for not only intrachromosomal deletions and duplications but also small supernumerary marker chromosomes 15, sSMC (15) [14]. Most sSMC (15) s take the form of a dicentric inv dup and can be classified into two groups: small sSMC (15) s and large sSMC (15) s. The small sSMC (15) s are metacentric chromosomes without euchromatic material and do not contain the Prader-Willi/Angelman Critic Region (PWACR), which usually clinically irrelevant. In contrast, the large sSMC (15) s are acrocentric chromosomes containing copies of PWACR and are frequently associated with abnormal phenotypes [15,16]. Consistently, in our case, the duplication was limited to 15q11.1-q11.2 and there were only two copies and combined no other aberrations (Figures 2 and 3). It is also important to point out that this duplication is de novo. Both parents have a normal karyotype. Clinical outcome may different between the familial case and the de novo case. From literature, 76% of the familial cases are without clinical signs while 75% of the reported de novo cases show clinical abnormalities. However, from studies performed in new-born cohorts, that de novo sSMC are associated with clinical signs only in 11-30% [17].
Conclusion
We conclude that array CGH is a modern and precise diagnostic tool that will complement and enhance current methods of detecting chromosomal imbalances prenatally. In combination of a detailed ultrasound examination and karyotype analysis, it can provide more precise and rapid prenatal diagnosis of sSMC.
Consents
Written informed consent was obtained from the patient for publication of this case report and any accompanying images. The study was accepted by the Ethics Committee of the hospital and they encourage publishing the article. A copy of the written consent is available for review by the Editor-in- Chief of this journal.
References
- Liehr T. Characterization of prenatally assessed de novo small supernumerary marker chromosomes by molecular cytogenetics. Methods Mol Biol 2008.; 444: 27-38.
- Liehr T. Handling small supernumerary marker chromosomes in prenatal diagnostics. Expert Rev Mol Diagn 2009; 9: 317-324.
- Liehr T, Weise A. Frequency of small supernumerary marker chromosomes in prenatal, newborn, developmentally retarded and infertility diagnostics. Int J Mol Med 2007; 19: 719-731.
- Viersbach R, Engels H, Gamerdinger U, Hansmann M. Delineation of supernumerary marker chromosomes in 38 patients. Am J Med Genet 1998; 76: 351-358.
- Warburton D. De novo balanced chromosome rearrangements and extra marker chromosomes identified at prenatal diagnosis: clinical significance and distribution of breakpoints. Am J Hum Genet 1991; 49: 995-1013.
- Chen CP, Lin CC, Ko TM, Tsai FJ, Chern SR. Prenatal diagnosis and molecular cytogenetic characterization of a small supernumerary marker chromosome derived from chromosome 21. Taiwan J Obstet Gynecol 2010; 49: 377-380.
- Bo WYX, Jieping S, Weipeng W, Yanping T. Potential speciation in humans involving Robertsonian translocations. Biomed Res 2013.
- Chien SC, Chen CP, Lin CC, Huang LC, Hsieh CT. Prenatal diagnosis of mos45,X/46,X,+mar in a fetus with normal male external genitalia and a literature review. Taiwan J Obstet Gynecol 2009; 48: 292-295.
- Pietrzak J. Molecular cytogenetic characterization of eight small supernumerary marker chromosomes originating from chromosomes 2, 4, 8, 18, and 21 in three patients. J Appl Genet 2007; 48: 167-175.
- ACOG Committee Opinion No. 446: array comparative genomic hybridization in prenatal diagnosis. Obstet Gynecol 2009; 114: 1161-1163.
- Wang T, Duan C, Shen C, Xiang J, He Q. Detection of complex deletions in chromosomes 13 and 21 in a fetus by noninvasive prenatal testing. Mol Cytogenet 2016; 9: 3.
- Shuqin X, Kun F, Yanzhi X, Jieping S, Weipeng W, Maowei C, Bo W. Analysis of meiotic segregation patterns and inter-chromosomal effects in sperm from a Robertsonian translocation family. Biomed Res 2014; 25: 233-239.
- Chen CP. Prenatal diagnosis and molecular cytogenetic characterization of mosaicism for a small supernumerary marker chromosome derived from ring chromosome 4. Taiwan J Obstet Gynecol 2011; 50: 188-195.
- Horsthemke B, Wagstaff J. Mechanisms of imprinting of the Prader-Willi/Angelman region. Am J Med Genet A 2008; 146: 2041-2052.
- Eggermann K, Mau UA, Bujdoso G, Koltai E, Engels H. Supernumerary marker chromosomes derived from chromosome 15: analysis of 32 new cases. Clin Genet 2002; 62: 89-93.
- Kleefstra T, de Leeuw N, Wolf R, Nillesen WM, Schobers G. Phenotypic spectrum of 20 novel patients with molecularly defined supernumerary marker chromosomes 15 and a review of the literature. Am J Med Genet A 2010; 152: 2221-2229.
- Liehr T. Familial small supernumerary marker chromosomes are predominantly inherited via the maternal line. Genet Med 2006; 8: 459-462.