Research Article - Research and Reports on Genetics (2018) Volume 2, Issue 1
Preliminary assessment of genetic diversity at the haemoglobin locus in the Bunaji cattle.
Ukwu HO*, Gwaza DS, Apav PM, Eruh SD
Department of Animal Breeding and Physiology, College of Animal Science, University of Agriculture, PMB 2373 Makurdi, Benue State, Nigeria
- *Corresponding Author:
- Ukwu HO
Department of Animal Breeding and Physiology
University of Agriculture
Nigeria
Tel: 234445332045
E-mail: ukwuhenry@gmail.com
Accepted Date: October 26, 2017
Citation: Ukwu HO, Gwaza DS, Apav PM, et al. Preliminary assessment of genetic diversity at the haemoglobin locus in the Bunaji cattle. J Res Rep Genet. 2017;1(1):25-28
Abstract
This study was designed to ascertain the level of genetic diversity at the haemoglobin (Hb) locus in the Bunaji cattle of Nigeria. Haemoglobin genotyping was performed on Seventy Bunaji cattle in Makurdi, Nigeria, using cellulose acetate electrophoresis. Data on Haemoglobin genotypes were subjected to chi-square test. Three haemoglobin genotypes (HbAA, HbAB and HbBB) controlled by two co-dominant haemoglobin alleles HbA and HbB were observed. The genotype frequencies were 0.50, 0.23 and 0.27 for HbAA, HbAB and HbBB respectively; genotype frequencies at haemoglobin locus in the Bunaji cattle were not in Hardy-Weinberg equilibrium. The gene frequencies of HbA and HbB were 0.61 and 0.39 respectively, with HbA being the most frequent. There was gene-controlled diversity at the haemoglobin locus in the Bunaji cattle with heterozygosity (He) value of 0.47, which is an indication of moderate level of genetic diversity at the haemoglobin locus in the Bunaji cattle in Makurdi, Nigeria.
Keywords
Bunaji Cattle, Diversity, Genotype, Hemoglobin locus, Heterozygosity.
Abbreviations
Hb: Hemoglobin; df: Degree of Freedom; N: Sample size; HWE; Hardy-Weinberg Equilibrium; NaCl: Sodium Chloride; EDTA: Ethylene-Diamine-Tetra-Acetic Acid; He: Heterozygosity
Introduction
The Bunaji cattle are the most numerous and widely spread of all the Nigerian cattle breeds; representing about 14.73 million cattle consisting of 1.47 million milking cows and 13.26 million beef cattle [1]. The Bunaji cattle is a tropically-adapted breed and beef animal, but its potential as a dairy animal has not been adequately harnessed, although it is kept mostly by its owner for milk. It can be identified by its white coat color, with black colour at the body extremities. In Nigeria, the Bunaji cattle are mostly kept by nomadic Fulani men who trek long distance with their herds from Northern region to Southern region of Nigeria yearly in search of pasture. Hence, in response to natural selection i.e., survival of the fittest, some of the Bunaji cattle die during trekking. Consequently, the Bunaji cattle in Nigeria may have evolved adaptabilities for its survivability under such conditions of hard trekking as well as response environmental influence. However, the Bunaji cattle of Nigeria has not been fully exploited for its genetic potentials using molecular and biochemical approach. Polymorphisms occurring at protein level could be used for preliminary study of the Bunaji cattle in developing country like Nigeria where state-of-the-art laboratory facilities for high throughput molecular genetic analysis are lacking.
Haemoglobin is a blood protein responsible for transport of oxygen and carbon (iv) oxide in the blood of vertebrates. Haemoglobin has been one of the most studied polymorphisms in vertebrate species since the infancy of both the population and evolutionary genetics [2]. This blood protein has been reported to exhibit polymorphism at its globin portion, but is known to be the same in its “haem portion” in all vertebrates [3]. The use of haemoglobin polymorphism in genetic analysis of farm livestock species has been tried out. For instance, an investigation of hemoglobin polymorphism in Ogaden cattle in Somali region of Southeastern Ethiopia revealed three haemoglobin genotypes HbAA, HbAB and HbBB, with HbA as the most prevalent allele in the population. The polymorphism of haemoglobin in cattle has been reported to be breed influenced as some breeds show a clear polymorphism with two alleles while others present only one allele [4]. The occurrence of individual cattle characterized by the presence of a novel α-globin variant, whose primary structure differs from the normal counterparts for the p.Ala27Th substitution in Agerolese cattle of Southern Italy has been reported by Salzano et al. [5]. In addition, polymorphism at the hemoglobin locus had been investigated in Nigerian indigenous chickens [6-8] in ducks [9,10] and in sheep [11].
Although the existence of three hemoglobin genotypes HbAA, HbAB and HbBB in the Bunaji cattle in Zaria, Nigeria had been reported by Essien et al. [12] there is dearth of information with respect to polymorphism at hemoglobin locus in the Bunaji cattle found in other ecological locations in Nigeria. Thus, there is need for further investigation with respect to the populations of the Bunaji cattle found in other parts of Nigeria. This study therefore was designed to assess within-breed genetic diversity at the haemoglobin locus in the Bunaji cattle in Makurdi, Nigeria.
Materials and Methods
Location of study
The research was conducted in Makurdi area of Benue state, Nigeria. Benue state lies within the lower river Benue trough in middle belt region of Nigeria. It geographic coordinate are longitude 70 471 and 100 01 East; latitude 60 251 and 80 81 North. The area is characterized by a period of dry season from October to March, and a period of rainy season from April to September. Annual rainfall ranges from 973 mm to 1324 mm.
Blood sample collection
Blood samples were collected from the jugular vein of 70 adult, healthy Bunaji cattle in Makurdi, Benue State, Nigeria. Blood samples were collected following the international guidelines for ethical treatment of animals in Applied Animal Behaviour and Welfare Research as prepared by the International Society for Applied Ethology (ISAE) Ethics Committee, 2002. Blood samples were drawn separately into vacationers (SARSTEDT Monovette®) containing Ethylene-diamine-tetra-acetic acid (EDTA) as anticoagulant, using vacationer needles. The samples were properly labeled according to the sex of the cattle. Haemoglobin genotyping of samples was performed using the facility at TOSEMA diagnostic laboratory, located in high level Makurdi, Benue state.
Hemoglobin genotyping
Hemoglobin genotypes were determined using cellulose acetate electrophoresis in a Shandon electrophoresis tank. About 0.5 ml of whole blood was placed into a centrifuge tube and spun for 30 minutes. 10 mL of cold 0.155 molar solution of NaCl was added to wash the red cells. About 4 volumes of Hb- Genotype lysing fluid was added to 1 volume of saline washed packed red cell in a clean test tube, mixing and leaving for 20 minutes. Equal volumes of Tris-EDTA-borate buffer with pH 8.5-8.6 were placed to the anode and cathode compartment of the electrophoresis tank. Cellulose acetate strips (77 × 150 mm) were prepared and labeled properly. They were soaked in Tris EDTA-Boric acid buffer (TEB) at a pH of 8.6 and blotted slightly with a filter paper to remove excess buffer. 5 ml of each of the haemolysate samples (tests and controls) was transferred into the well plate, carefully applying the haemolysate samples including controls on a slightly blotted strip using an applicator. The strip was placed on a bridge and then lowered into the compartments containing the buffer in the electrophoresis tank. It was allowed to run at 300 V for 30 minutes. After completion of the electrophoresis run, the haemoglobin pattern, in order of motility was detected based on the molecular weight of the hemoglobin molecules HbAA, HbAB and HbBB directly without drying or staining, by noting the position, number and intensity of the bands on the strip. The direct gene counting method was used to score Hb bands based on the separation of Hb variants. Human control hemoglobin (Hb-AA and Hb-AS) were used to develop bovine haemoglobin control for the 70 blood samples drawn from the Bunaji cattle. The experiment was repeated and result used as comparable variant to the observed haemoglobin control bands on the cellulose acetate strip.
Statistical analysis
Genotype frequencies of the haemoglobin genotypes and allele frequencies of the Hb alleles were estimated. Genotype frequencies were calculated as follows:
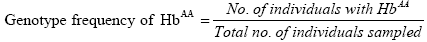
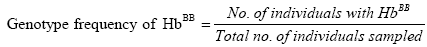
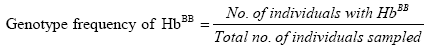
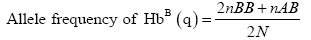
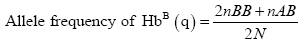
Where:
nAA=Number of individuals with HbAA genotype.
nAB=Number of individuals with HbAB genotype.
nBB=Number of individuals with HbBB genotype.
N=Total number of individuals sampled.
Data on genotype frequencies were subjected to Chi-square analysis to test for goodness-of-fit for observed and expected frequencies under Hardy-Weinberg equilibrium (HWE). Yate’s correction for continuity was performed during Chi-square test because expected frequencies between 5 and 10 were observed, and only one degree of freedom was observed. Estimates of Heterozygosity were calculated as shown by the expression below [13].
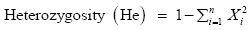
Where: n=Number of loci
Xi=Frequencies of the alleles.
Results
The genotype and allele frequencies at the hemoglobin locus in Bunaji cattle are shown in Table 1. The observed and expected numbers of hemoglobin genotypes in Bunaji bulls are shown in Table 2. The observed and expected numbers of hemoglobin genotypes in Bunaji cows are shown in Table 3, while Table 4 shows the observed and expected numbers of haemoglobin genotypes in the pooled (bull and cow) samples of Bunaji cattle.
Table 1. Genotype and allele frequencies at the haemoglobin locus in the Bunaji cattle in Makurdi, Nigeria.
| Sex |
N | Genotype number | Genotype frequencies | Allele frequencies | |||||
|---|---|---|---|---|---|---|---|---|---|
| HbAA | HbAB |
HbBB | HbAA |
HbAB | HbBB | HbA | HbB | ||
| Male | 33 | 18 | 7 | 8 | 0.55 | 0.21 | 0.24 | 0.655 | 0.345 |
| Female | 37 | 17 | 9 | 11 | 0.46 | 0.24 | 0.30 | 0.580 | 0.420 |
| Total | 70 | 35 | 16 | 19 | 0.50 | 0.23 | 0.27 | 0.614 | 0.386 |
Table 2. Observed and expected number of haemoglobin genotypes in the Bunaji Bulls.
| Hb genotypes | Observed | Expected | X2 df=1 |
|---|---|---|---|
| HbAA | 18 | 14.158 | |
| HbAB | 7 | 14.914 | |
| HbBB | 8 | 3.928 | 8.783** |
**(P<0.01)
Table 3. Observed and expected number of haemoglobin genotypes in the Bunaji Cows.
| Hb genotypes | Observed | Expected | X2 df=1 |
|---|---|---|---|
| HbAA | 17 | 12.447 | |
| HbAB | 9 | 18.026 | |
| HbBB | 11 | 6.527 | 8.772* |
*(P<0.01)
Table 4. Observed and expected number of haemoglobin genotypes in the Bunaji Cattle (Pooled – bulls and cows).
| Hb genotypes | Observed | Expected | X2 df=1 |
|---|---|---|---|
| HbAA | 35 | 26.390 | |
| HbAB | 16 | 33.181 | |
| HbBB | 19 | 10.430 | 18.158*** |
***(P<0.001)
Three distinct hemoglobin genotypes HbAA, HbAB and HbBB, which are controlled by two co-dominant alleles HbA and HbB were observed in this study. The haemoglobin genotype frequencies observed in this study were 0.55 (HbAA), 0.21 (HbAB) and 0.24 (HbBB) in Bunaji bulls, and 0.46 (HbAA), 0.24 (HbAB) and 0.30 (HbBB) in Bunaji cows. The most frequent haemoglobin genotype observed in this study was HbAA with genotype frequency of 0.50, while the genotype frequencies of HbAB and HbBB were 0.23 and 0.27, respectively, in the pooled samples of the Bunaji cattle in Makurdi, Nigeria. The gene (allele) frequencies of HbA and HbB observed in this study were 0.61 and 0.39, respectively.
The results of Chi-square test were significant Tables 2-4. This implies that the observed and expected genotype frequencies at the haemoglobin locus in Bunaji cattle in Makurdi were not in Hardy-Weinberg proportion.
The estimated heterozygosities at the hemoglobin locus in Bunaji cattle are shown in Table 5. The estimated heterozygosity in the entire (Pooled) population was 0.47.
Table 5. Heterozygosities at haemoglobin locus in the Bunaji cattle in Makurdi.
| Groups | Heterozygosities |
|---|---|
| Bulls | 0.45 |
| Cows | 0.49 |
| Entire (pooled) population | 0.47 |
Discussion
Several haemoglobin alleles exist and this has led to the appearance of different haemoglobin genotypes in farm livestock. The diversity at haemoglobin locus could confer advantage to individuals bearing it, or may be detrimental to individuals bearing it. The hemoglobin locus in the Bunaji cattle in Makurdi, Nigeria is polymorphic with three distinct genotypes – HbAA, HbAB and HbBB controlled by two alleles. The three hemoglobin genotypes observed in this study were the same with the observation of Essien et al. [12] in Bunaji cattle in Zaria, Kaduna state. The gene frequencies of HbA (0.614) and HbB (0.386) observed in this study were comparable with the results of Essien et al. [12] who reported gene frequencies of 0.64 (HbA) and 0.36 (HbB), with slight differences. Similar haemoglobin genotypes were also reported in Ogaden cattle found in Southeastern part of Ethiopia with genotype frequencies of 54.2% (HbAA), 33.3% (HbAB) and 12.3% (HbBB), and gene frequencies of 0.709 (HbA) and 0.291 (HbB) [14]. The preponderance of HbA allele as shown in this study and previous report [14] could be an indication that the presence of HbA allele could confer a degree of natural selective advantage to individuals bearing it in terms of survivability in their natural environment.
The result of chi-square showed that genotype frequencies at haemoglobin locus in the Bunaji cattle were not in Hardy- Weinberg equilibrium. The reason could be due to the sample size, or forces that affect gene and hence genotype frequency in a population (e.g., natural or artificial selection). The disequilibrium of genotype frequency at the haemoglobin locus in the Bunaji cattle observed in this study is not in agreement with the earlier observation in the Bunaji cattle [12] and Ogaden cattle [14]. This could be due to the management system of keeping the herds. The samples used in this study were collected from several open herds, while samples used in previous studies [12,14] were taken from presumably closed herds maintained in research institute and beef farm. Undoubtedly, equilibrium of gene and hence genotype frequencies is assured after one generation of random mating with Hardy-Weinberg condition brought under control in such closed populations.
Heterozygosity is a measure of genetic diversity or variability in a population. The heterozygosity value of 0.47 observed in this study is an index of moderate genetic diversity at the hemoglobin locus in Bunaji cattle in Makurdi. Genetic differences and similarities within and between breeds are important raw materials for genetic improvement of animals [4]. The observed heterozygosity at the haemoglobin locus in the Bunaji cattle is beneficial since genetic diversity within a population enables perpetuation of the species in the presence of changing climatic conditions. This shows clearly that haemoglobin locus in the Bunaji cattle in Nigeria is not controlled by only one allele.
Conclusion
The results of this study suggest that the haemoglobin locus in the Bunaji cattle in Makurdi, Nigeria, is controlled by two codominant alleles HbA and HbB. The three haemoglobin genotypes observed in this study were HbAA, HbAB and HbBB. The allele HbA was the most frequent Hb allele in the population of the Bunaji cattle studied. There exists a moderate genetic diversity at the haemoglobin locus in the Bunaji cattle in Makurdi, middle belt of Nigeria. Further research should be carried out to determine the effect of the observed haemoglobin polymorphism on productive performance of the Bunaji cattle in Nigeria.
Acknowledgement
We wish to thank the management and staff of TOSEMA Laboratory, High Level, Makurdi, Benue State, for the facilities used in Haemoglobin genotyping.
References
- Tibi KN, Aphunu A. Analysis of the cattle market in Delta state - The supply determinants. African Journal of General Agriculture. 2010;6(4):199-203.
- Pieragostini E, Alloggio I, Petazzi F. Insights into Haemoglobin Polymorphism and Related Functional Effects on Haematological Pattern in Mediterranean Cattle, Goat and Sheep. Diversity. 2010;2(4):679-700.
- Chineke CA, Ologun AG, Ikeobi CON 2007. Haemoglobin Types and Production Traits in Rabbit Breeds and Crosses. J Biological Sci. 2007;7(1):210-4.
- Egena SSS, Alao RO.2014. Haemoglobin Polymorphism in Selected Farm Animals- A Review. Biotechnology in Animal Husbandry. 2014;30(3):377-90.
- Salzano AM, Pauciullo A, D’Ambrosio C, et al. Bovine hemoglobin polymorphism: a novel alpha-globin variant identified in the Agerolese breed from southern Italy. Czech J Anim Sci. 2015;60(4):145-51.
- Salako AE, Ige AO. Haemoglobin polymorphisms in Nigerian indigenous chickens. JAnim Vet Adv. 2006; 5(11):897-900.
- Yakubu A, Aya VE. Analysis of genetic variation in normal feathered, naked neck and Fulani-ecotype Nigerian indigenous chickens based on haemoglobin polymorphism. Biotechnol Anim Husb. 2012;28(2):377-84.
- Ajayi FO, Agaviezor BO, Wihioka SN. Haemoglobin genotypes in the Nigerian indigenous chicken in the Niger Delta region of Nigeria. Int J Adv Biol Res. 2013;3(1):13-6.
- Akinyemi MO, Osaiyuwu OH, Ajayi AY. Biochemical characterization of Muscovy and Mallard Ducks in Nigeria. Int J Sci and Nat. 2014;5(3):557-62.
- Oguntunji AO, Ayorinde KL. Blood protein polymorphism and genetic diversity in locally adapted Muscovy duck (Cairina moschata) in Nigeria. Animal Genetic Resources. 2015;1-10.
- Akinyemi MO, Salako AE. Hemoglobin Polymorphism and Morphometrical Correlates in the West African Dwarf Sheep of Nigeria. Int J Morphol. 2010;28(1):205-8.
- Essien IC, Akpa GN, Barje PP, et al. Haemoglobin types in the Bunaji cattle and their Friesian crosses in Shika, Zaria-Nigeria. African J Anim Biochemistry. 2011; 6(1):112-6.
- Nei, M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583-90.
- Pal SK, Mummed YY. Investigation of haemoglobin polymorphism in Ogaden Cattle. Veterinary World. 2014;7(4):229-33.