Research Article - Journal of Molecular Medicine and Therapy (2017) Journal of Molecular Medicine and Therapy (Special Issue 1-2017)
Position dependence of an enhancer activity of the human beta-globin intron-ii, within a heterologous gene.
- *Corresponding Author:
- Alireza Zomorodipour
Department of Molecular Medicine National Institute of Genetic Engineering and Biotechnology Tehran
Tel: 203 974 7892
E-mail: dawn.foster@yale.edu
Accepted date: October 10, 2017
Citation: Zomorodipour A, Jahromi EM, Ataei F, et al. Position dependence of an enhancer activity of the human beta-globin intron-ii, within a heterologous gene. J Mol Med Ther 2017;1(1):19-24.
Abstract
Function of intron II of the human beta globin on the human factor IX (hFIX) expression, with respect to its position within the coding region, was investigated. With this aim two hFIX expressing plasmids, in which the intron was inserted in either second or 3'-terminal intronic positions of the cDNA, in parallel with an intron-less clone, were used for the hFIX expression analysis in two mammalian cell lines, in transient states. Protein assays demonstrated the highest hFIX expression level for the construct with intron in the second position. This was also evidenced by a 3-fold increase in the hFIX transcription, relative to the variant with intron in the terminal position. The recombinant hFIX expressed by the three minigenes were biologically active, as it was demonstrated by clotting test. Results presented in this study, support the position dependence of an intron regulatory function, in an exogenous state, by effecting at either transcriptional or/and posttranscriptional level.
Keywords
Hemophilia-B, Human coagulation factor IX (hFIX), Human β -globin (hBG)- introns, Intronic position effect, Mammalian expression system.
Introduction
Hemophilia B, an X-linked recessive bleeding disorder, is caused by the functional deficiency or lack of coagulation factor IX (FIX) [1]. FIX is an essential vitamin K-dependent (VKD) protein that participates in the intrinsic pathway of blood coagulation [2]. Currently, replacement therapy is the major treatment for hemophilia B, carried out via infusion of normal hFIX, produced from either human plasma or recombinant mammalian expression systems [3].
Presence of introns within the transcribed sequences and in the proper orientation in their normal locations, provides gene structure, similar to normal mammalian genes [4,5]. Experimental evidences showed that either partial or full length of an intron, may improve the expression of its linked gene [6,7]. Truncated forms of the hFIX first intron, increased the hFIX expression up to nine-folds, in cultured hepatoma cells [8]. In transgenic mice, inclusion of the full-length 6.2-kb or a truncated 1.4-kb fragment of the hFIX intron-I increased the gene expression up to 200-fold [9]. Export of the hBG transcripts is highly dependent on the presence of its native introns, in particular the second one [10]. A number of groups reported improvements of transgene expressions, by in vivo applications of the hBG intron-II [8,11]. There are reports indicating that introns in their native positions and in different positions yield opposite outcomes of gene expression [12,13]. More related to present work is the inclusion of either of the hBG introns I or II, that could restore the expression of an intron-less rabbit β -globin gene in HeLa cells [13]. Insertion of the hBG intron I into the corresponding location in dehydrofolate reductase (Dhfr) cDNA improved the production of Dhfr even more than the time when the Dhfr native intron I was applied, in vitro [7]. In similar studies, applications of the first and second introns of the rabbit β -globin in the human factor VIII and ceruloplasmin cDNAs successfully improved the productions of their corresponding proteins [14]. In our previous study the enhancer-like activity of the hBG intron II inside the coding region of the hFIX on its expression level, in cultured mammalian cell, was demonstrated [15]. A significant increase in the hFIX expression level occurred whenever the hBG-intron II was present in the minigene. As a step forward, in this work we have studied the function of the hBG intron II, with regards to its position, on the hFIX expression. With this aim, a recombinant CMV-regulated hFIX expressing plasmid was constructed in which the hBG intron II was inserted in the 3'-terminal intronic position of the hFIX cDNA. The recombinant plasmid in parallel with two other hFIX expressing plasmids, reported previously [15], including; a plasmid with the hBG intron II in second intronic position and an intron-less one, were used to transfect two mammalian cell lines. Since the complete hBG intronic sequences were inserted in the hFIX exon junction sites in the constructed plasmids, the corresponding minigenes were expected to contain requirements for proper splicing. Subsequently, the transfected cells were used to study transient expressions of the hFIX in a comparative manner.
Materials and Methods
Bacterial strain, mammalian cell line, plasmids and primers
The DH5 α strain of Escherichia coli was used as host for various cloning and subcloning steps. The human embryonic kidney (HEK-293T) and Chinese hamster ovary (CHO) cell lines (obtained from Pasteur Institute, Iran) were cultured and transfected as described previously [15]. Plasmid pBCE392 [15] was used as template for amplifications of the hBG intron II and as the hFIX cDNA source. Plasmid pcDNA3 (Invitrogen-USA) was used for the construction of the hFIXexpressing plasmid. Two CMV-regulated hFIX expressing plasmids, pcDNA3-hFIX (named F9 in this work; with an intron-less hFIX cDNA) and pcDNA3-hFIX-intII-2 (named F9INT2 in this work; having the hBG intron II in the second intronic position of the hFIX cDNA) were described previously [15]. Oligonucleotides, used for PCRs and RT-PCRs (listed in Table 1) were synthesized by MWG-Germany. To facilitate cloning steps, restriction sites were considered at the 5' ends of the designed oligonucleotides.
| Name | Oligonucleotide Sequences | Application |
|---|---|---|
| hF9KozF | GGATCCGCCACCATGCAGCGCGTGAACATGAT | PCR |
| emj R1 | CGAGCTCCTGCAGTTACAATCCATTTTTC | PCR |
| hIX-R1 | GCGGCCGCAGTGATTAGTTAGTGAGAGGCCC | PCR |
| emj F2 | AAAACTGCAGCCCACTGTGTTGAAACTGGTGTTAAAAT | PCR |
| TACAGTTGTCGCAGGTGAGTCTATGGGACGCTTGATG | ||
| emj R3 | CAATATTATGTTCACCTGTGGGAGGAAGATAAG | PCR |
| emj F4 | CTTATCTTCCTCCCACAGGTGAACATAATATTG | PCR |
| Hu-gapdhF1 | AAGGTGAAGGTCGGAGTCAAC | RT- PCR |
| Hu-gapdhR2 | GGGGTCATTGATGGCAACAATA | RT- PCR |
| hF9-RTF1 | GGTGGTTTGCTCCTGTACTGA | RT- PCR |
| hF9-RTR1 | CGGGTGAGCTTAGAAGTTTGTG | RT- PCR |
Table 1: Details of satellite imageries used.
Molecular techniques
All DNA manipulations were carried out based on standard cloning procedures [16]. A plasmid, carrying the hBG intron II in the last intronic position of the hFIX cDNA, was constructed by inserting the intron within the acceptor-donor site between the exons 7 and 8 of hFIX-cDNA in three steps. In the first step, oligonucleotides hFIX-Koz and emjR1 were used to amplify an 836 bp fragment covering the hFIX exons 1 to 7 (E1-7). This fragment, carrying BamHI and PstI/SacI sites on its 5' and 3' ends respectively, was subcloned between BamHI and SacI sites in a pET26 plasmid, generating plasmid pET26- E1-7. To insert the intron in the terminal intronic position, a Spliced-Overlap Extension PCR approach was considered. First, oligonucleotides emjF2 and emjR3 were used to amplify the hBG intron II carrying a section of 3' end of the hFIX exons 7 on its 5' end (with PstI) and a part of the 5' end of the hFIX exon 8 on its 3'-end. Secondly, oligonucleotides emjF4 and hIX-R1 were used to generate a second fragment covering the hFIX exon 8, with a piece of the 3'-end of the hBG intron II on its 5'-end and a NotI site on its 3' end. The two PCR products were joined to create E7-intII E8, a chimeric fragment carrying PstI and NotI sites on its 5' and 3' ends, respectively. The PCR product was subsequently inserted next to exon 7 in pET-E1-7, from previous step, to generate pET-F9INT7. In the third step, the hFIX mini-gene having the hBG intron-II (between Exons 7 and 8), was excised from pET-F9INT7 after BmaHI/NotI digestion and subcloned into a pcDNA3 plasmid to generate plasmid F9INT7.
Online sequence analysis
The hFIX and hBG nucleotide sequences were obtained from GenBank (NCBI) with the accession numbers of AY769950 and L48217 respectively [17].
Cell culture and transfection
The mammalian cells were grown in Dulbeco's modified Eagle's medium (DMEM) and Hams-F12 (Gibco-BRL Life Technology, Germany) at a 1 to 1 ratio supplemented with 10% (v/v) heat inactivated fetal bovine serum (FBS; Gibco-BRL Life Technology, Germany) plus 100 U/mL of penicillin G and 100 μ g/mL of streptomycin (Sigma, Germany). The cells were transfected with approximately 2 μg plasmid DNA based on a lipofection-based method, using FuGene-6. The transfection reaction continued for up to 7 hours. Subsequently, the cultured medium was harvested and a fresh rich medium (containing Vitamin K and FBS) was added. 7 hours after transfection, conditioned medium was replaced by fresh medium containing 1 β -g vitamin K1/ml. On the 2nd day of post-transfection, the cultured medium was collected for transient expression analysis and fresh medium was added.
Measurement of the hFIX expression
Biological activity of the secreted hFIX was measured using immuno-depleted plasma for FIX and activated partial thromboplastin reagent (aPTT; cephalite) (with a sensitivity limit of 0.01 U/ml), according to the manufacturer's instructions (Diagnostica Stago- France). The FIX::Ag in the conditioned cultured media was assayed by ELISA, as described previously [10], according to the manufacturers' instructions (Asserachrom, Diagnostica Stago-France).
RNA preparation and reverse transcription PCR (RTPCR)
Using a kit for RNA isolation (Cinnagen-Iran), total RNAs were extracted from the cells, following the manufacturer’s procedure. Using the isolated RNA, cDNA was synthesized by reverse transcriptase (M-MuLV; Roche-Germany) and random hexamer and used for amplification of a piece of the hFIX cDNA, with two hFIX-specific primers, hF9-RTF1 and hF9- RTR1 (Table 1). In order to investigate the occurrence of the intron splicing, the prepared cDNA was subsequently analyzed by the amplification of a piece of the hFIX cDNA, using oligonucleotides, hF9KozF and hIX-R1 (Table 1).
Quantitative real time RT-PCR (Q-RTPCR)
To investigate the considerable variability of FIX expression due to the intron influence, Q-RT-PCR on the RNA prepared in the previous step, and the comparative ΔCT method [18] were applied, using gene-specific SYBR Green-based Quanti-Tect® Primer assays (QIAGEN, Germany). The Q-RT-PCR data was analyzed based on the comparative threshold (Ct) method, and the fold expressions of samples were compared with that of an intron-less clone during 72h of post-transfection. The human GAPDH was used as an internal reference gene to normalize the expression of the transgene.
Statistical analysis
Protein expression analysis including coagulation test and ELISA, were carried out in triplicates and the generated data were presented as the means ± SD. The data collected were subjected to analysis of variance test. The means were compared using Duncan’s multiple range tests. A p-value of less than 0.05 was considered as significant difference. All statistical analyses were carried out with SPSS 11.5 (SPSS Inc.).
Results
Transient expression of hFIX
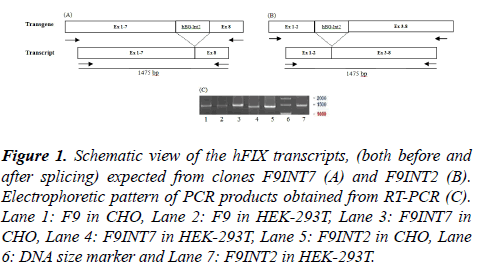
Analysis of the hFIX transcript: Two different mammalian cell lines, CHO and HEK293T, were separately transfected transiently with the three recombinant plasmids namely; F9, F9INT2 and F9INT7. Forty eight hours after transfections, the hFIX transcripts were evaluated by performing RT-PCR, using hF9KozF/hIX-R1 primer pair. Based on the primers' positions, PCR products of an appropriate size with 1475bp length was expected from both the intron-less and intron containing minigenes (Figure 1).
Figure 1: Schematic view of the hFIX transcripts, (both before and after splicing) expected from clones F9INT7 (A) and F9INT2 (B). Electrophoretic pattern of PCR products obtained from RT-PCR (C).Lane 1: F9 in CHO, Lane 2: F9 in HEK-293T, Lane 3: F9INT7 in CHO, Lane 4: F9INT7 in HEK-293T, Lane 5: F9INT2 in CHO, Lane 6: DNA size marker and Lane 7: F9INT2 in HEK-293T.
As the results shows, products of expected size were produced for all the examined cells that is in consistent with proper and complete splicing of pre-mRNAs from the intron-containing constructs, which is supposed to be equal to its intron-less counterpart. Use of oligonucleotides emjF2 and emjR3 (specifically designed for the hBG intron II) on the same templates did not lead to any product (not shown).
In order to evaluate the possible function of the intron at transcription level, Q-RT-PCR was performed. Based on the results obtained from the quantification of the hFIX transcripts, about 0.6- and 2-fold(s) increased for the mRNA contents of the F9INT7 and F9INT2 clones, respectively, compared with the parental clone (Figure 2).
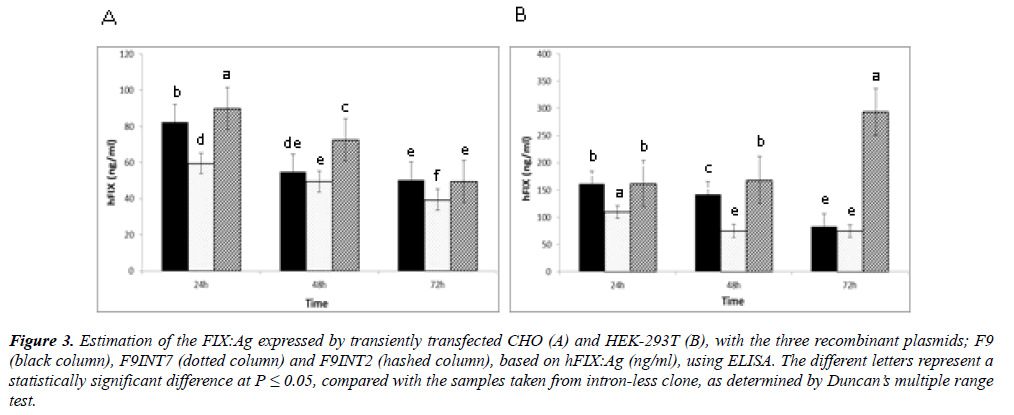
Measurement of the hFIX::Ag: For any of the transfectants, presence of hFIX::Ag in the cultured media was examined by performing ELISA during 72 hours of post-transfection. As the data from the measurements of the FIX::Ag indicated, the highest hFIX expression for each of the cell lines were obtained, when they were transfected with F9INT2 (Figure 3). In contrast, the cells with F9INT7, showed the lowest hFIX expression levels in similar conditions, in both of the cell lines. Although during this period the hFIX expression variations of the two cell lines were different, the HEK293T cells, with the expression level of about 300 ng/ml/106 cells, appeared more efficient for the expression of FIX, after 72 hours of post transfection. Whereas, for the CHO cells the highest FIX expression level, with about 90 ng/ml, occurred 24 hours after transfection.
Figure 3: Estimation of the FIX:Ag expressed by transiently transfected CHO (A) and HEK-293T (B), with the three recombinant plasmids; F9 (black column), F9INT7 (dotted column) and F9INT2 (hashed column), based on hFIX:Ag (ng/ml), using ELISA. The different letters represent a statistically significant difference at P < 0.05, compared with the samples taken from intron-less clone, as determined by Duncan?s multiple range test.
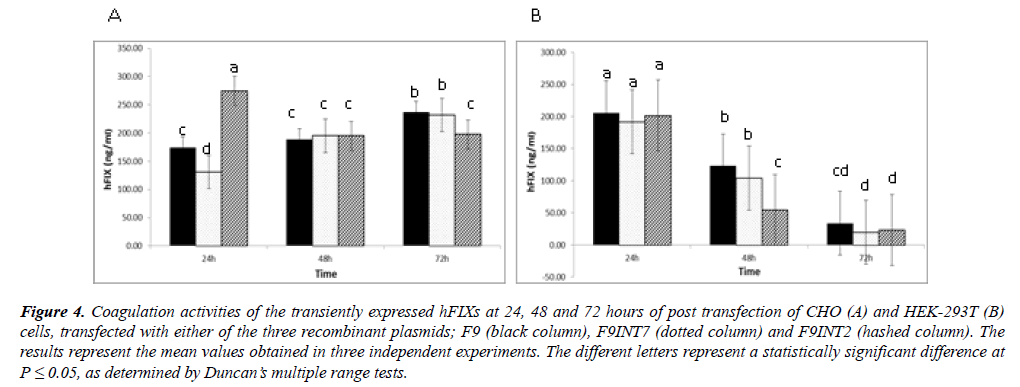
Activity measurements of the expressed rhFIX: The cultured media taken from the transfected cells were used for FIX clotting activity measurements, at 24, 48 and 72 hours of post-transfection (Figure 4). In general, the mean coagulation activities of the cultured media taken from the three examined constructs were significantly higher than that of the negative control, indicating in the expression of biologically active FIX by the examined recombinant cells. A sharp decrease in the coagulation activities of cultured media taken from the HEK-293 cells during 72 hours of post-transfection was evidenced. Whereas, constant trends of coagulation activities were observable in the cases of the CHO cultures, during the same period.
Figure 4: Coagulation activities of the transiently expressed hFIXs at 24, 48 and 72 hours of post transfection of CHO (A) and HEK-293T (B) cells, transfected with either of the three recombinant plasmids; F9 (black column), F9INT7 (dotted column) and F9INT2 (hashed column). The results represent the mean values obtained in three independent experiments. The different letters represent a statistically significant difference at P = 0.05, as determined by Duncan?s multiple range tests.
Discussion
A number of independent works demonstrated that both the human and rabbit β-globin introns play critical roles in the expression of the linked gene, when they are inserted within coding regions [7,13,14] In our previous study the enhancerlike activity of the hBG introns I and II inside the coding region on the rhFIX expression level, in cultured mammalian cells, was demonstrated [15]. In those experiments, significant increase in the hFIX expression level occurred whenever the hBG-intron II was present in the construct. Therefore, involvement of factors with enhancer-like activities in this intron was speculated. Such activities could occur due to elements like TFB sites, acting through their promoter proximity positions that is supported by a number of potential TFBs in the hBG-intron II [18]. Presence of a 96 bp sequence, which belongs to the L1 family of non-LTR retrotransposons (NLRs), within the second hBG intron may also contribute to the enhancer-like activity of this intron as it has already been suggested by Le Hir et al. [19].
Introns also take part in gene regulation through splicing, polyadenylation (PA), nuclear mRNA transport, RNA editing, RNA silencing, mRNA stability, mRNA decay, localization and translational efficiency of mRNA [20-23]. A growing number of examples reveal an interconnection between these processes and intron splicing [23-25]. To address the position-dependence of the hBG intron II regulatory role, it was examined in two intronic positions of the hFIX cDNA. First, the proper removal of the hBG intron II in both positions from pre-mRNAs of the transfected cells, was demonstrated by RTPCR.
As our quantitative protein assays indicated, the highest hFIX expression level, for each of the cell lines, occurred for the F9INT2 minigene. In accordance with our protein results, about 3-fold increase in the transcript level of hFIX, due to the presence of the hBG-derived intron in the second intronic position occurred, relative to the case with the intron in the terminal position. Presence of introns, were shown to be crucial for the accumulation of mRNA in certain genes, such as growth hormone [8] and purine nucleoside phosphorylase [23]. The results obtained in this work, is in agreement with this idea that, an expression level increase, mediated by intron, might occur at the level of mRNA accumulation, as it has been pointed out by other groups [26,27]. The function of hBG intron II on its native gene expression has been correlated to its position in the hBG gene as the last intron, where it is essential for the accumulation of stable cytoplasmic mRNA and is implicated in promoting efficient 3'-end formation [10]. The necessity of a functional 3'-terminal intron for an efficient 3'- end formation of transcript was also emphasized by studies carried out on the human glycolytic enzyme triosephosphate isomerase (TPI) [28]. Some elements within the 3'-end intron have been shown in a number of genes, which interact with polyadenylation processing and increase the poly-A tail length, leading to longer half-life for mRNA and more efficient translation [29]. The relatively low expression level of the F9INT7 mini-gene, which carries an exogenous terminal intron in a terminal intronic position, is not in consistent with the above-mentioned states. However, the relatively higher hFIX expression level of F9INT2 mini-gene than that of the intron less F9 may support the advantage of a functional intron with its inherent intronic features [7,8,21].
Harding et al. [11] used the second hBG intron located between a liver-specific promoter and the hFIX cDNA and showed a significant increase in the hFIX expression level after addition of the hBG 2nd intron. In another work insertion of the hBG intron II between the mouse metallothionein I promoter and the rat growth hormone cDNA, caused a substantial increase at the mRNA level in vivo [8]. Improvement of transgene expression by applying the hBG-derived intron has also been shown by other investigators [14]. The result presented in this paper is in principle in agreement with the findings of the abovementioned investigators. However, there is a major difference between this work and those of others, in which the major results were obtained from in vivo analysis. The result obtained from an in vitro experiment (cell culture) is usually different from those of in vivo studies [11] and the results obtained from an in vitro system with that of in vivo system may not be comparable.
As our quantitative expression data indicates, the higher expression of the hFIX in the HEK-293T cells in comparison with that in CHO cells in similar conditions was evidenced, that could be explained by stable expression of the large Tantigen of SV40 in the HEK-293T cells, causing the effective replication of episomal plasmids [30]. However, based on the results obtained from clotting tests the HEK-derived FIX appeared to be less stable during 72 hours of post transfection, when compared with that of the CHO cells.
Conclusion
The results presented in this paper are from transient expression state that provided evidences in favor of the hBGderived intron enhancer-like activity, which affects with respect to its position within the linked gene. The evidences provided at both transcript and protein levels, by effecting at either transcriptional or/and posttranscriptional level(s). Next step to this work is to examine the function of the hBG intron II in different intronic positions in stable transfection states.
Acknowledgement
This work was financially supported by a grant (No: 375) from the National Institute of Genetic Engineering and Biotechnology of Iran.
References
- Tuddenham E, Schwaab R, Seehafer J, et al. Haemophilia A database of nucleotide substitutions deletions insertions and rearrangements of the factor VIII gene. Nucleic Acids Res. 1994;22(22):4851-68.
- Jackson CM, Nemerson Y. Blood coagulation. Annu Rev Biochem. 1980;49:765-811.
- Hedner U, Ginsburg D, Lusher JM, et al. Congenital hemorrhagic disorders: new insights into the pathophysiology and treatment of hemophilia. Hematology Am Soc Hematol Educ Program. 2000:241-65.
- Donath M, Mendel R, Cerff R, et al. Intron-dependent transient expression of the maize GapA1 gene. Plant Mol Biol. 1995;28(4):667-76.
- Lacy-Hulbert A, Thomas R, Li X, et al. Interruption of coding sequences by heterologous introns can enhance the functional expression of recombinant genes. Gene Ther. 2001;8(8):649-53.
- Lu S, Cullen BR. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA. 2003;9(5):618-30.
- Noé V, MacKenzie S. An intron is required for dihydrofolate reductase protein stability. J Biol Chem. 2003;278(40):38292-300.
- Palmiter RD, Sandgren EP, Avarbock MR, et al. Heterologous introns can enhance expression of transgenes in mice. Proc Natl Acad Sci U S A. 1991;88(2):478-82.
- Jallat S, Perraud F, Dalemans W, et al. Characterization of recombinant human factor IX expressed in transgenic mice and in derived trans-immortalized hepatic cell lines. EMBO J. 1990;9(10):3295-301.
- Antoniou M, Geraghty F, Hurst J, et al. Efficient 3-end formation of human Ã-globin mRNA in vivo requires sequences within the last intron but occurs independently of the splicing reaction. Nucleic Acids Res. 1998;26(3):721-9.
- Harding T, Koprivnikar K, Tu G, et al. Intravenous administration of an AAV-2 vector for the expression of factor IX in mice and a dog model of hemophilia B. Gene Ther. 2004;11(2):204-13.
- Bourdon V, Harvey A, Lonsdale DM. Introns and their positions affect the translational activity of mRNA in plant cells. EMBO Rep. 2001;2(5):394-8.
- Buchman AR, Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol. 1988;8(10):4395-405.
- III C, Yang C, Bidlingmaier S, et al. Optimization of the human factor VIII complementary DNA expression plasmid for gene therapy of hemophilia A. Blood Coagul Fibrinolysis. 1997;8 Suppl 2:S23-30.
- Haddad MA, Zomorodipour A, Izadpanah M, et al. A systematic study of the function of the human Ã-globin introns on the expression of the human coagulation factor IX in cultured Chinese hamster ovary cells. J Gene Med. 2009;11(10):941-50.
- Benson DA, Karsch MI, Lipman DJ, et al. GenBank. Nucleic Acids Res. 2006;34:D16-20.
- Sambrook J and Russell DW. Molecular cloning: a laboratory manual (3-volume set);2001.
- Haddad MZA, Hosseini SJ, Sabouni F. In silico investigation and application of human beta-globin introns as hetrologous elements to increase the expression of human coagulation Factor IX. Iran J Biol. 2009;11:2 98-107
- Le Hir H, Nott A, Moore MJ. How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci. 2003;28(4):215-20.
- Fong YW, Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001 20-27;414(6866):929-33.
- Nott A, Meislin SH, Moore MJ. A quantitative analysis of intron effects on mammalian gene expression. RNA. 2003;9(5):607-17.
- Wiegand HL, Lu S, Cullen BR. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc Natl Acad Sci USA. 2003;100(20):11327-32.
- Jonsson JJ, Foresman MD, Wilson N, et al. Intron requirement for expression of the human purine nucleoside phosphorylase gene. Nucleic Acids Res. 1992;20(12):3191-8.
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416(6880):499-506.
- SträÃer K, Hurt E. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature. 2001;413(6856):648-52.
- Rethmeier N, Seurinck J, Montagu M, et al. Intron-mediated enhancement of transgene expression in maize is a nuclear, gene-dependent process. Plant J. 1997;12(4):895-9.
- Rose AB, Last RL. Introns act post-transcriptionally to increase expression of the Arabidopsis thaliana tryptophan pathway gene PAT1. Plant J. 1997;11(3):455-64.
- Nesic D, Cheng J, Maquat LE. Sequences within the last intron function in RNA 3'-end formation in cultured cells. Mol Cell Biol. 1993;13(6):3359-69.
- Furger A, Sullivan JMO, Binnie A, et al. Promoter proximal splice sites enhance transcription. Genes Dev. 2002;16(21):2792-9.
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23(1):175-82.