Research Article - Biomedical Research (2018) Volume 29, Issue 18
Population genetic characteristic of horses of Mugalzhar breed by STR-markers
Seleuova LA*, Naimanov DK, Jaworski Z, Aubakirov MZH, Mustafin MK, Mustafin BM, Safronova OS, Baktybaev GT, Turabaev AT and Domatski VN
Baitursynov State University, Kostanay, Baytursynov Str., 47, Kazakhstan
Accepted date: September 26, 2018
DOI: 10.4066/biomedicalresearch.29-18-1041
Visit for more related articles at Biomedical ResearchAbstract
One of the most important tasks of improvement of horse breeds is investigation into genetic determinants responsible for formation of high productivity and use of genetic monitoring upon selection control. Nowadays genome selection is the most advanced estimation method of breeding abilities. This article presents results of genetic testing of DNA of Mugalzhar horses by 16 microsatellite loci, from 5 to 13 alleles have been identified. Genetic diversity (Nv) of modern population of purebred brood horses was at the level of 7.882, polymorphism (??) equalled to 4.457, heterozygosity (??)-to 0.754, ??-to 0.775, fixation index (Fis)-to 0.027; it referred to a cluster of native/local breeds. Polymorphism varied from 2.785 to 7.442; actually observed heterozygosity from 0.138 to 0.931; theoretically expected one-from 0.641 to 0.866. The number of active alleles in population (Na) was equal to 134.0.
Keywords
Genome selection, Horse breed, Molecular genetic markers, Polymorphism, Population, Primer
Introduction
Monitoring of population genetic polymorphism is one of the most important tasks of preservation and reproduction of various agricultural breeds including horses [1,2]. In this regard characteristics of population genetic structure can be the first step to breed preservation and reproduction, in addition it improves cattle breeding [3,4].
Such analysis is carried out on using molecular genetic technologies based on various DNA markers [3,5]. This approach expands significantly capabilities of population genetic analysis, makes it possible to reveal cross- and inbreed variety of separate genome segments, presents breed genetic structure and serves as excellent tool for investigation into genetic variance in the frames of genus or species [5-10].
In 1998 year (Order No. 156 of Ministry of Agriculture of Kazakhstan dated December 30, 1998) the researchers and horse breeders of Kazakhstan, using pure breeding and improvement of Jabe horse breed, created specialized dualpurpose Mugalzhar horse breed which could provide high quality meat and kumis during yearlong grassland farming [11,12].
Taking into account high value of the domestic Mugalzhar horse breed and increased demand for meat and dairy products of horse breeding, it would be reasonable to preserve and to improve the Mugalzhar breed in Kazakhstan on a wider range, this stipulated the targets of our work.
Practical value of the studies: the obtained results evidence activity of genetic processes upon horse breeding in «Sholak Espe» farm, high polymorphism and high genetic diversity of the considered horse breed. The available data on horse genotypes make it possible to perform more efficient selection aimed at preservation of genetic peculiarities of lines (family groups) and support of inbreed differentiation.
Materials and Methods
The studies were performed with samples of biological material of 70 Mugalzhar horses bred in «Sholak Espe» farm in Karaganda region (Central Kazakhstan). We used hair bulbs of the tested horses as biological material for DNA isolation.
These studies were performed in test laboratory of TOO «Kazak Tulpary» (Ministry of agriculture of Kazakhstan, Kostanay) certified according to ISO/IEC 17025-2009.
Genome DNA was isolated from hair bulbs of horses with proper modifications using reagents produced by OOO «Izogen», Russia, according to the producer recommendations. Genetic typing of the horses was carried out using a set of 16 microsatellite loci recommended by International Society for Animal Genetics (ISAG): HVL20, HTG4, AHT4, HMS7, HTG6, AHT5, HMS6, ASB23, ASB2, HTG10, HTG7, HMS3, HMS2, ASB17, LEX3, HMS1. The amplification products were identified using an ABI Prism 310 (Applied Biosystems, USA) genetic analyzer on the basis of capillary electrophoresis and laser detection. The acquired graphical results were processed using Gene Mapper 4.0 software. Polymorphism was characterized by the following indices: allele frequency of the considered loci calculated by maximum likelihood formula (pA → Equation (1)) and genotype frequency (PAA → Equation (2)), actually observed (Ho → Equation (3)) and theoretically expected heterozygosity (He → Equation (4)) with consideration for the Hardy-Weinberg law Ho-according to Ney, He (with Ca according to Robertson → Equation (5)), as well as average heterozygosity for populations, fixation index (Fis → Equation (6)), polymorphism level (Ae → Equation (7)), average number of alleles in locus (Nv → Equation (8)).
pA=(2 nAA+nAB+nAC+)/2N → (1)
where 2 nАA was the double number of homozygotes; n АВ, n АС were the number of heterozygotes; 2N was the double number of analysed animals in the selection [13].
pAA=nAA/N → (2)
where р АА was the y frequency of genotypes of the considered DNA microsatellite loci; n AA was the number of animals with genotype АА [13].
H0=hj/n → (3)
where Ho was the actually observed heterozygosity for one locus; hj was the number of heterozygote genotypes in locus; n was the total number of genotypes in locus [14].
He=1-Ca → (4)
where He was the expected heterozygosity; Са was the expected homozygosity, it was determined on the basis of coefficient of homozygosity by the Robertson formula [15].
Ca=Σnp2 → (5)
where Са was the expected homozygosity; p was the gene frequency of alleles; n was the number of alleles in locus [15].
Fis=1-(Ho /He ) → (6)
where Ho was the actually observed heterozygosity; He was the expected heterozygosity [15].
Ae=1/(Σpij2 ) → (7)
where Ае was the level of polymorphism (active effective alleles); р was the occurrence frequency of the jth allele for locus I, and summation was applied for n alleles [14].
Na=1/Ca → (8)
where Na was the number of active alleles in population (polymorphism); Ca was the level of expected homozygosity of horses by polymorphic alleles [13].
Results and Discussion
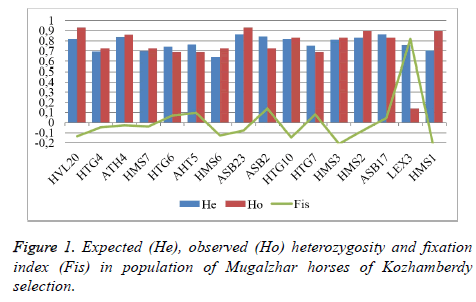
While analyzing the allele pool of the considered horses by 16 DNA microsatellites, we obtained data characterizing polymorphism of each marker (Table 1).
| Microsatellite locus | Expected homozygosity (Са) | Polymorphism (Ае) | Observed heterozygosity (Но) | Expected heterozygosity (Не) | Fixation index (Fis) | Number of alleles (Na) |
|---|---|---|---|---|---|---|
| HVL20 | 0.182 | 5.497 | 0.931 | 0.818 | -0.138 | 11 |
| HTG4 | 0.308 | 3.247 | 0.724 | 0.692 | -0.046 | 6 |
| AHT4 | 0.162 | 6.184 | 0.862 | 0.838 | -0.028 | 8 |
| HMS7 | 0.303 | 3.298 | 0.724 | 0.697 | -0.039 | 7 |
| HTG6 | 0.260 | 3.849 | 0.690 | 0.740 | 0.068 | 6 |
| AHT5 | 0.235 | 4.247 | 0.690 | 0.765 | 0.098 | 6 |
| HMS6 | 0.359 | 2.785 | 0.724 | 0.641 | -0.130 | 7 |
| ASB23 | 0.137 | 7.313 | 0.931 | 0.863 | -0.079 | 11 |
| ASB2 | 0.160 | 6.253 | 0.724 | 0.840 | 0.138 | 10 |
| HTG10 | 0.184 | 5.426 | 0.828 | 0.816 | -0.015 | 10 |
| HTG7 | 0.249 | 4.014 | 0.690 | 0.751 | 0.082 | 5 |
| HMS3 | 0.189 | 5.289 | 0.828 | 0.811 | -0.021 | 7 |
| HMS2 | 0.170 | 5.881 | 0.897 | 0.830 | -0.080 | 8 |
| ASB17 | 0.134 | 7.442 | 0.828 | 0.866 | 0.044 | 13 |
| LEX3 | 0.240 | 4.163 | 0.138 | 0.760 | 0.818 | 7 |
| HMS1 | 0.293 | 3.412 | 0.897 | 0.707 | -0.268 | 5 |
| Average | 0.2243 | 4.4574 | 0.7546 | 0.7757 | 0.0272 | 134.0 |
Table 1. Polymorphism of the considered microsatellite loci of DNA of Mugalzhar horses.
On the basis of the considered 16 STR-loci from 5 to 13 alleles were identified. According to Table 1, it was established that average polymorphism of locus (Ае) was 4.457, hence, all loci were subdivided into two groups. The first group was comprised of loci with the level of polymorphism below average: HMS6, HTG4, HMS7, HMS1, HTG6, HTG7, LEX3, AHT5. Minimum value was that of HMS6: 2.785. The second group was comprised of loci with the level of polymorphism above average: ASB17, ASB23, ASB2, AHT4, HMS2, HVL20, HTG10, HMS3. Maximum polymorphism was that of ASB17: 7.442.
In order to obtain precise estimation of population variance, the index of expected heterozygosity was introduced which considered the level of allele diversity. In this regard we estimated the actually observed and the theoretically expected heterozygosity calculated by 16 loci, the respective average indices were 0.7546 and 0.7757.
The highest actually observed heterozygosity (Но) was that of locus HVL20 (0.931), and that of theoretically expected heterozygosity (Не)-locus ASB17 (0.866), whereas the minimum heterozygosity was that of locus LEX3 (0.138), located on sex chromosome, and the minimum expected heterozygosity was that of locus HMS6 (0.641).
The calculated fixation index Fis made it possible to establish interrelation among the considered horses of Mugalzhar breed of Kozhamberdy selection. We revealed that the deficiency of heterozygotes was observed for loci HTG6, AHT5, ASB2, HTG7, ASB17, LEX3 (Figure 1).
For preservation of genetic inbreed diversity, average number of alleles (Nv) for all the considered markers in the breed were of great concern. In the Mugalzhar breed it was 7.882.
In addition, we counted the number of active alleles in population by Na, which also characterized the level of polymorphism. This variable was inversely proportional to the Robertson homozygosity coefficient. The highest level of polymorphism was observed for the Mugalzhar horses on locus ASB17 [13]. This variable in the aggregate for all considered loci amounted to 134.0.
While studying published information, we revealed that the method of determination of genetic polymorphism of horses and other farm livestock using marker microsatellites was widely applied in practice. Thus, Chasymova et al. while estimating genetic variation in populations of Tuvinian horses, detected from 4 to 9 alleles in the considered microsatellite loci, in our study this parameter was from 5 to 13 alleles. Microsatellite loci ASB17, АНТ4, VHL20, ASB2 and ASB23 were presented by the most varied range of alleles which had been confirmed by our results. However, in Mugalzhar horses the loci HMS2, HMS3 were also marked as loci of high polymorphism [16].
Mahrous et al. reported about high information content of micorsatellite loci AHT4, HTG10, ABS2, ABS23 which also corresponded with our experimental results [17].
It is possible to state that application of microsatellite markers has high resolution ability making it possible to estimate consolidarity of the considered stock, to perform more efficient selection of animals, to control inheritance of valuable properties.
Conclusion
On the basis of the considered 16 STR-loci from 5 to 13 alleles were identified. The average polymorphism of locus (Ае) was 4.457, hence, all loci were subdivided into two groups. The first group was comprised of loci with the level of polymorphism below average: HMS6, HTG4, HMS7, HMS1, HTG6, HTG7, LEX3, AHT5. Minimum value was that of HMS6: 2.785. The second group was comprised of loci with the level of polymorphism above average: ASB17, ASB23, ASB2, AHT4, HMS2, HVL20, HTG10, HMS3. Maximum polymorphism was that of ASB17: 7.442. The actually observed heterozygosity was from 0.138 to 0.931; the theoretically expected one was from 0.641 to 0.866, average indices were 0.7546 and 0.7757, respectively.
The highest actually observed heterozygosity (Но) was that of locus HVL20 (0.931), and that of theoretically expected heterozygosity (Не)-locus ASB17 (0.866), whereas the minimum heterozygosity was that of locus LEX3 (0.138), located on sex chromosome, and the minimum expected heterozygosity was that of locus HMS6 (0.641).
The calculated fixation index Fis made it possible to establish interrelation among the considered horses of Mugalzhar breed of Kozhamberdy selection. We revealed that the deficiency of heterozygotes was observed for loci HTG6, AHT5, ASB2, HTG7, ASB17, LEX3. Average number of alleles (Nv) for all considered markers in Mugalzhar horse breed was 7.882.
In addition, we counted the number of active alleles in population by Na, which also characterized the level of polymorphism. This variable was inversely proportional to the Robertson homozygosity coefficient. The highest level of polymorphism was observed for the Mugalzhar horses on locus ASB17 [13]. This variable in the aggregate for all considered loci amounted to 134.0.
References
- Barcaccia G, Felicetti M, Galla G, Capomaccio S, Supplizi AV. Molecular analysis of genetic diversity, population structure and inbreeding level of the Italian Lipizzan horse. Livestock Science 2013; 151: 124-133.
- Metlitska OI. The Methodology of DNA gene pools certification of farm animals for hypervariable loci in the genome. Poltava 2012.
- Kalashnikov VV, Zaitsev AM, Khrabrova LA. Genetic identification and origin monitoring in horse breeding. Scientific support of development and efficiency of brood, sports and productive horse breeding in Russia and CIS countries: Proceedings of International Conference devoted to the 75th anniversary of Prof. Koveshnikov, 2014.
- Suprun IА. Evaluation of phylogenetic connections in liverstock breeding. Golovna 2014.
- Rothschild MF, Plastow GS. Applications of genomics to improve livestock in the developing world. Livestock Science 2014; 166: 76-83.
- Ganopoulos I, Kalivas A, Kavroulakis N, Xanthopoulou A, Mastrogianni A, Koubouris G, Madesis P. Genetic diversity of Barbary fig (Opuntia ficus-indica) collection in Greece with ISSR molecular markers. Plant Gene 2015; 2: 29.
- Korbin M, Kuras A, Zurawicz E. Fruit plant germplasm characterisation using molecular markers generated in RAPD and ISSR-PCR. Cell Mol Biol Lett 2002; 7: 785-794.
- Sarla N, Bobba S, Siddiq EA. ISSR and SSR markers based on AG and GA repeats delineate geographically diverse Oryza nivara accessions and reveal rare alleles. India Curr Sci 2003; 84: 683.
- Stolpovskiy YA. Application inter-SSR DNA analysis to assess population structure, identification and similarity of gene pools of species and breeds of domesticated animals. Genetics 2010; 46: 825-833.
- Zabek T, Nogay A, Radko A, Nogay J, Slota E. Genetic variation of Polish endangered Bilgoraj horses and two common horse breeds in microsatellite loci. Appl Gene 2005; 46: 299-305.
- Satybaldin AA. Current state of horse breeding and horse sports in Kazakhstan. The First International Conference Kostanay 2002.
- Turabaev A. New lines in Kulandy interbreed type of Mugalzhar horse breed. Novosti Nauki Kazakhstana 2011; 156.
- Estimation procedure of genetic diversity and genetic similarity of farm and local breeds. Divovo 2011.
- Kantanen J. Genetic diversity and population structure of 20 north European cattle breeds. J Heredity 2000; 91: 446-457.
- Methodology of application of DNA analysis of horses for estimation of genetic resources in horse breeding. Divovo 2011.
- Chasymova RB, Khrabrova LA, Zaitsev AM, Makarova EYu, Fedorov YuN, Ludu BM. Estimation of genetic variety in populations of Tuvinian horses according to loci of blood system and microsatellite DNA. Sel`skokhozyaistvennaya Biologiya 2017; 52: 679-685.
- Mahrous KF, Hassanane M, Mordy A, Shafey HI, Hassan N. Genetic variations in horse using microsatellite markers. J Genetic Eng Biotechnol 2011; 9: 103-109.