Research Article - Biomedical Research (2017) Volume 28, Issue 5
Plasmid transfer capacities of multi- resistant UPEC clinical isolates in biofilms
Yan Qing Tong1, Min Sun1, ChunJie Hu2, DongKai Zhao2*1Department of Nephrology, the Affiliated Hospital to Changchun University of Chinese Medicine, Changchun City, Jilin Province, PR China
2Department of Microbiology, the Affiliated Hospital to Changchun University of Chinese Medicine, Changchun City, Jilin Province, PR China
- *Corresponding Author:
- DongKai Zhao
Department of Microbiology
The Affiliated Hospital to Changchun
University of Chinese Medicine, PR China
Accepted on October 31, 2016
Abstract
Uropathogenic Escherichia coli (UPEC) is the leading causative agent of Urinary tract infection (UTI). The emerging multidrugresistance of UPEC to antimicrobial agents (such as antibiotics or Chinese herbs) has been a serious public health problem and may result in the complication or failure of UTI therapy, yet with unknown underlying mechanisms. Our present study aims to explore the plasmid DNA-based gene transfer mechanism of the drug resistance UPEC strains. The pPICZαA plasmid containing the Zeocin resistance gene was applied as extraneous resistance plasmid to investigate the uptake ability of plasmid and the plasmid transfer frequency of a UPEC stain alone or co-culturing with DH5α bacteria during the formation of biofilm process. The results showed that the uptake efficiency and plasmid transfer frequency increased from 8 h to 24 h during the biofilms formation, and then slowed down after the formation of biofilms. What’s more, the plasmid transfer frequency was independent of the bacteria culture conditions. Our present study is of importance to the mechanisms illustration of multidrug resistance (like herbal medicine) in UPEC, and might provide a foundation for the research of resistance gene spreading reduction.
Keywords
Uropathogenic Escherichia coli (UPEC); Urinary tract infection (UTI); Multidrug-resistance; Biofilms; Resistance gene spreading.
Introduction
Urinary tract infection (UTI) refers to the infection in any part of urinary system [1]. It is one of the most common types of nosocomial infection in humans, and about 150 million new UTI cases were reported annually all over the world. Among all the predominant microorganism or etiologic agent that causes UTIs, Uropathogenic Escherichia coli (UPEC) is proved to be the major bacterial culprit cause (about 80%) [2-4].
The UPEC causing UTI makes it one of the most common indications for conventional antimicrobial therapy. As a result, the excessive use of antibiotics leads to emergence and rapid increase in antibiotic resistance in UPEC. Recently studies indicated that the rise of multidrug resistance (MDR) in UPEC leaded to the aggregation and complication of UTI disease, such as a higher rate of recurrent, protracted course of UTI, which eventually could result in parenchymal destruction or renal insufficiency of patients [3,5,6]. The emergence and increasing prevalence of MDR in UPEC has now been a serious public health problem. Thus it is of vital importance to explore the virulence mechanisms of MDR-UPEC prevalence for developing new generation of therapeutic agents.
Chinese herbal medicine or extract was proved to be novel therapy method for UTI treatment. However, the application of Chinese herb extract for UTI therapy also leads persistent exposure of UPEC to herb extract therapeutic drugs, and eventually results in the rise of Chinese herb extract-resistant UPEC isolates [7]. Recently, our group successfully isolated some UPEC strains clinically from UTI patients and it all showed various resistance to a Chinese herbal extracts (data not shown). The strains could expressed pga genes, which were discovered to be responsible for the synthesis and export of polysaccharide β-1,6-N-acetyl-D-glucosamine polymer (PGA), which was the material of biofilm formation, and luxS genes, which encoded auto-inducer 2 (AI-2) acting as a signal molecule of quorum-sensing system for the biofilm formation. We hypothesized that drug resistance mechanism mediated by biofilm formation may play a vital role in the multidrug resistance of UPEC.
In present study we aimed to investigate the plasmid DNAbased gene transfer as well as the antibiotic resistances mechanisms of a Chinese herb extract-resistant UPEC strain (1351). The pPICZαA plasmid containing the Zeocin resistance gene was applied as extraneous resistance plasmid DNA to investigate the uptake of plasmid and the plasmid transfer of UPEC alone or co-culturing with DH5α bacteria during the formation of biofilm process.
Materials and Methods
Drug-resistance test of UPEC strains
Three clinically isolated UPEC strains 1351 was inoculated onto solid culture medium containing different concentrations of kana, chloramphenicol, tetracycline, zeocin and hygromycin, respectively. The concentrations of antibiotics were 16 μg/ml, 32 μg/ml, 64 μg/ml, 128 μg/ml, 256 μg/ml. According to the growth of bacterial colonies in the culture plate, the growth of the UPEC was determined. The resistance of strain was determined according to the standard of antimicrobial susceptibility testing.
Uptake of exogenous plasmids by UPEC strain
Monocolony of UPEC 1351 was seeded in 10 ml of LB culture medium and cultured for 18 h with shaking. The bacteria solution was diluted to 0.5 Mc Farland and then, 2 μl was taken and added into 96 well plates containing 200 μl medium dilution. To investigate the influence of exogenous plasmid concentration on UPEC strain uptakefinal concentration of 0.5 μg/ml, 1 μg/ml, 2 μg/ml, 4 μg/ml and 8 μg/ml pPICZA plasmids were incubation with the biofilms for 24 hours. The samples were diluted and seeded onto Zeocin-containing and free plates. The colonies were counted for evaluation of the uptake of exogenous antibiotics-resistant plasmid. Exogenous plasmid pPICZA was added at the final concentration of 2 μg/ml. To determine whether the intake of exogenous antibiotics-resistant plasmid by UPEC 1351 was timedependent, UPEC 1351 and exogenous plasmids pPICZA was co-incubated for 4 h, 8 h, 12 h, 16 h, 24, 36 and 48 h, respectively. At each time point, Samples were taken from three wells and incubated in 37°C for 24 h. Then, the samples were diluted and coated onto Zeocin-containing and free plates. The colonies were counted.
To determine the difference between strains with biofilm and those without biofilm, equal exogenous plasmids were added into the two different strains. After 24 h and 48 h incubation, the intake of exogenous plasmids was measured as described above. Plasmid delivery during mutualism culture and biofilm formation of UPEC and DH5 α (pPICZA+). To determine the optimal observation time for drug-resistant plasmid transfer during mutualism culture, the mutualism culture of DH5 (pPICZA+) and UPEC single colony were firstly performed in LB medium (pH7.4). The transfer frequency of drug-resistant plasmids was measured using the above described method after bacteria seeding 4 h, 8 h, 12 h, 16 h, 24 h, 36 h and 48 h, respectively. Briefly, co-cultured bacteria were coated onto plates containing both Zeocin and UPEC1351-resistant herb extract drug. The grown colonies were counted.
Subsequently, 0.5% sugar or 100 mM CaCl2 containing LB and TSB medium at the pH of 7.4 (normal), 6.0 (partial acid) and 9.0 (alkaline) were used for mutualism culture of DH5α (pPICZA+) and UPEC single colony. At the optimal observation time as determined above, plasmid delivery frequencies in different medium were measured using the above method.
Statistics
Data was expressed as Mean ± SD. Statistical analysis was performed using SPSS 17.0. A p<0.05 was considered as statistically significant.
Results
Drug-resistance of UPEC strain (1351)
The UPEC strain 1351 could all grow well in culture plates with 16 μg/ml - 256 μg/ml of antibiotics kanamycin sulfate, chloromycetin and tetracycline. But the growth was all inhibited in culture plates with 16 μg/ml - 256 μg/ml of both Zeocin and Hygromycin (Table 1). The results suggested that the isolated UPEC strain 1351 was sensitive to Zeocin and Hygromycin. Accordingly, the pPICZαA plasmid contains the Zeocin resistance gene was used as extraneous resistance plasmid DNA for plasmid uptake and transfer investigation.
| Antibiotic | Kana | Chloramphenicol | Tetracycline | Zeocin | Hygromycin |
|---|---|---|---|---|---|
| 16μg/ml | + | + | + | - | - |
| 32μg/ml | + | + | + | - | - |
| 64μg/ml | + | + | + | - | - |
| 128μg/ml | + | + | + | - | - |
| 256μg/ml | + | + | + | - | - |
Illustration: “+” stands for positive, “-” stands for negative
Table 1. Measurement of drug-resistance for different antibiotics.
Influence of exogenous plasmid concentration on UPEC strain uptake
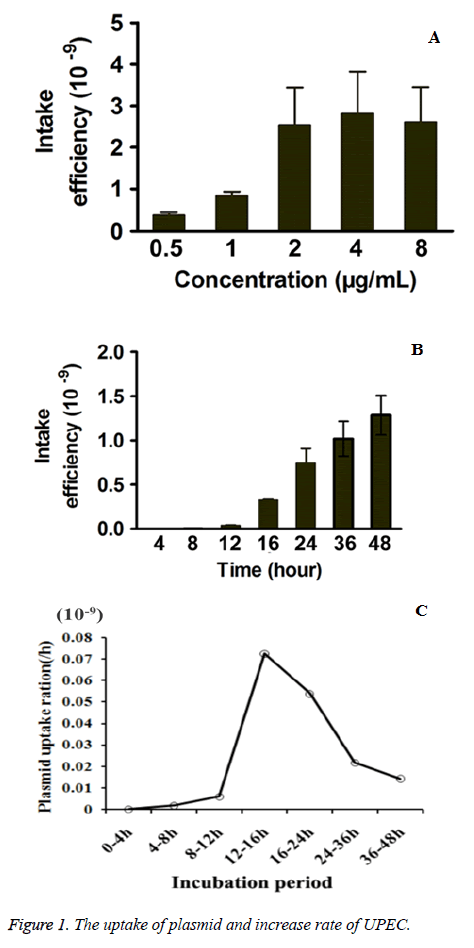
To further investigate the influence of exogenous plasmid concentration on UPEC strain uptake, 0.5 μg/ml, 1 μg/ml, 2 μg/ml, 4 μg/ml and 8 μg/ml of exogenous drug-resistant pPICZA plasmids were administrated to UPEC strain. As shown in Figure 1A, the plasmids uptake frequency increased with a dose-dependent manner from 0.5 μg/ml - 2 μg/ml, but exhibited no obvious increase over 2 μg/ml. Thus 2 μg/ml of plasmid was applied for further investigation.
Time-dependent uptake of plasmids by UPEC strain and influence of plasmid concentration
As shown in Figure 1B, the pPICZA plasmids uptake frequency of UPEC strain increase over culture time and was of time dependent manner. In order to further investigate the increase rate in the pPICZA plasmids uptake frequency, the increase of plasmids uptake frequency over per two time points was calculated by subtracting the uptake frequency of one time point from the uptake frequency of the next time point. As shown in Figure 1C, uptake frequency increase rate increased between 8 h-24 h, and especially the increase in 12 h-16 h and 16 h-24 h exhibited the highest rate among all time period, during which the strain finished the bacterial adhesion and biofilms formation. After the formation of biofilms, the increase rates slowed down.
The uptake frequency of biofilms and planktonic bacteria
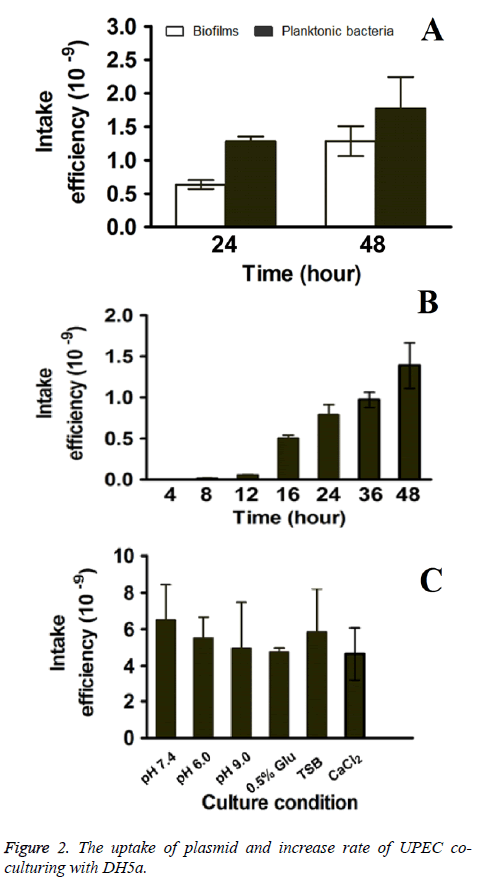
The uptake frequency of both biofilms and planktonic bacteria were evaluated to compare the uptake ability of multidrug resistant UPEC and normal E. coli. As shown in Figure 2A, the uptake frequency of E. coli in biofilms were 0.635 ± 0.041 × 10-9 and 1.284 ± 0.052 × 10-9 for 24 h and 48 h respectively, which it were lower than that of planktonic E. coli (1.281 ± 0.047 × 10-9 and 1.775 ± 0.057 × 10-9 for 24 h and 48 h respectively). The results indicated that exogenous drugresistant plasmids uptake frequency of planktonic bacteria was significantly high than that of UPEC in biofilms.
Influence of cultivation environments on the plasmid transfer capacities between UPEC and DH5α during biofilm formation
In order to study the plasmid transfer capacities between biofilms and planktonic bacteria during biofilm formation, UPEC and DH5α were co-cultured and the plasmid transfer was measured. Similarly with UPEC strains alone, the pPICZA plasmids uptake frequency of UPEC strain increase in a time dependent manner as co-culturing with planktonic bacteria (Figure 2B).
What’s more, the rate of increase in the pPICZA plasmids uptake frequency was also measured as mentioned above. The increase rates were 0 × 10-9, 0.0165 × 10-9, 0.0422 × 10-9, 0.449 × 10-9, 0.284 × 10-9, 0.1788 × 10-9, 0.4186 ×1 0-9 for 0 - 4 h, 4 h - 8 h, 8 h - 12 h, 12 h - 16 h, 16 h - 24 h, 24 h - 36 h, 36 h - 48 h respectively (Figure 2C), which indicated that the plasmid transfer capacities increase from 8 h to 24 h during the formation of biofilms and the increase rate slowed down after that. As increase rate exhibited the highest level (0.449 × 10-9) between 8 h - 12 h, the time point 12 h was further used for evaluation the influence of culture conditions on plasmids uptake frequency.
Further, different culture condition, the normal LB (pH 7.4) medium, acidic LB (pH 6.0) medium, alkaline LB (pH 9.0) medium, normal LB containing 0.5% glucose, TSB medium (pH 7.4) and normal LB containing 100 mM CaCl2 were applied. As shown in Figure 2C, there was no significant difference among all the groups. The results suggested that culture conditions (such as pH value and glucose concentration) had no influence on the plasmids uptake frequency.
Discussion
The emergence and rapid increase of MDR in UPEC made UTIs a serious public health problem and resulted in complicated therapy for it. In our previously study, we successfully isolated UPEC strains with various resistance to Chinese herbal extract clinically from UTI patients. The strains could expressed pga and luxS genes, which played important role during the formation and maturation of biofilm process. Our present study further confirmed that increase rate of uptake frequency of the UPEC strain increased along with the formation of biofilms from 8 h to 24 h and the increase rate was slowed down after biofilms formation. What’s more, the uptake frequency of UPEC strain was similar under different culture condition. As is well known, the bacterial biofilm refers to a structured community of bacterial cells as well as the selfproduced polymeric matrix around it [8-10]. The bacteria in biofilm are physiologically distinct from planktonic bacteria, which by contrast are single-cells floating or swimming in a liquid medium. The biofilms forming strains could exhibit obviously drug-resistance by reducing the antibiotic targets, such as the biosynthesis of bacterial walls, or reducing the antibiotic uptake [11]. The biofilms lead to the high drugresistance ability of bacteria, which may result in the evading of bacteria to the immune system, and eventually chronic infection as well as our control of diseases [12-15].
Recent studies indicated that the plasmid transfer may also occurred during the biofilms formation process, and the gene transfer was considered to be one of the major reasons for the obtainment of the drug-resistance genes [16,17]. Besides, the strains with antibiotic resistance genes could also spread the genes as the biofilms dissociation and caused more damage [18]. Thus the relationship between biofilms formation and gene transfer among strains is key to understanding mechanisms of drug-resistance strains emergence. In the present study, the Chinese herb extract-resistant UPEC strain was proved to uptake the exogenous drug-resistant plasmids pPICZA plasmids in a time dependent manners, and the increase rate of uptake frequency increased during biofilms formation process (8 h - 24 h), after which the increase rates slowed down. The bacteria of spatially structured bacterial populations or the biofilms are very different from planktonic bacteria in well-mixed liquid environments. The plasmid DNA-based gene transfer is favored by the bacterial biofilms due to the close cell-to-cell contact of the bacterial cells, since the densely packed biofilms cells could minimize shear force of it [19,20]. The biofilm was also proved to be beneficial for the increased stability of plasmid [21,22].
The development of biofilms involved in numbers of genes and proteins via many mechanisms. Firstly, several adhesin gene products were proved to mediate the development and competence of Escherichia coli biofilm, and biofilm associated adhesin genes could eventually enhance the ability of UPEC to cause UTI. For example, fimA encoding Type 1 fimbriae was proved to facilitate Escherichia coli’s attachment to the host mucosa as well as it biofilm formation. Evidences suggested that the expression levels of fimA varied in different time period, during which the biofilm formation abilities were different [23]. F9 genes (Such as gene clusters c1931-c1936 from UPEC CFT073), which encoded another type of fimbriae that were highly similar to those of type 1 fimbriae, could also strongly mediate biofilm growth of E. coli strain [24]. Secondly, some biofilm matrix related genes have been also proved to play important roles during biofilm formation.
The pga genes pgaABCD, which encoded the polysaccharide β-1,6-N-acetyl-D-glucosamine polymer (PGA) is the necessary element for biofilm formation [25,26]. Thirdly, the growth and metabolism associated genes or proteins could also mediate the development and competence of Escherichia coli biofilm, which may potentially affect the infection of biofilms. For example, as a part of the activated methyl cycle, LuxS is proved to affect biofilm formation through AI-2-independent effects on metabolism, which may assist it in adapting to diverse conditions and eventually lead to the high drugresistance ability [27]. What’s more, evidence proved that about 11 kinds of growth related link genes in biofilm were upregulated as compared to planktonic bacteria during logarithmic and stable phase. To sum up, the increase of biofilm matrix, or growth and metabolism associated genes expression contribute greatly to the biofilm development and thus were closed to the high drug-resistance ability of the bacteria. The biofilm lifestyle of bacteria which were mediated by these genes or proteins levels, lead to the gene transfer of genetic elements and increase the range of it, especially for the resistance genes under adverse environmental. Consistently, as demonstrated in our study, UPEC strain biofilm was proved to have an increased rate of transfer of a pPICZαA plasmid containing the Zeocin resistance gene compared to planktonic E. coli under zeocin condition.
Conclusion
The present study confirmed that persistent exposure of UPEC to herb extract therapeutic drugs could eventually result in the rise of Chinese herb extract-resistant UPEC isolates. Increase rate of the Chinese herb extract-resistant UPEC strain’s uptake frequency increased along with the formation of biofilms from 8 h to 24 h and the increase rate slowed down after biofilms formation. However, the difference of culture conditions had no significantly influence on the plasmids uptake frequency.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81374039).
References
- Niranjan V, Malini A. Antimicrobial resistance pattern in Escherichia coli causing urinary tract infection among inpatients. Indian J Med Res 2014; 139: 945-948.
- Derakhshandeh A, Firouzi R, Motamedifar M, Boroojeni AM, Bahadori M, Arabshahi S, Novinrooz A, Heidari S. Distribution of virulence genes and multiple drug-resistant patterns amongst different phylogenetic groups of uropathogenic Escherichia coli isolated from patients with urinary tract infection. Lett Appl Microbiol 2015; 60: 148-154.
- Hadifar S, Moghoofei M, Nematollahi S, Ramazanzadeh R, Sedighi M. Epidemiology of Multi Drug Resistant Uropathogenic Escherichia coli in Iran: a Systematic Review and Meta-analysis. Jpn J Infect Dis 2016.
- Justice SS, Hung C, Theriot JA, Fletcher DA, Anderson GG, Footer MJ, Hultgren SJ. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc Natl Acad Sci U S A 2004; 101: 1333-1338.
- Dehbanipour R, Rastaghi S, Sedighi M, Maleki N, Faghri J. High prevalence of multidrug-resistance uropathogenic Escherichia coli strains, Isfahan, Iran. J Nat Sci Biol Med 2016; 7: 22-26.
- Zhao R, Shi J, Shen Y, Li Y, Han Q, Zhang X, Gu G, Xu J. Phylogenetic Distribution of Virulence Genes Among ESBL-producing Uropathogenic Escherichia coli Isolated from Long-term Hospitalized Patients. J Clin Diagn Res 2015; 9: DC01-4.
- Tong Y, Xin B, Chi Y. Chinese herb-resistance and adherence to human uroepithelial cells of uropathogenic Escherichia coli. Afr J Tradit Complement Altern Med 2014; 11: 109-115.
- Chai Y, Chu F, Kolter R, Losick R. Bistability and biofilm formation in Bacillus subtilis. Mol Microbiol 2008; 67: 254-263.
- Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol 2010; 8: 623-633.
- Lebeaux D, Ghigo JM, Beloin C. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev 2014l; 78: 510-543.
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science 1999; 284: 1318-1322.
- Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov 2003; 2: 114-122.
- Donelli G, Guaglianone E. Emerging role of Enterococcus spp in catheter-related infections: biofilm formation and novel mechanisms of antibiotic resistance. J Vasc Access 2004; 5: 3-9.
- O'Connell HA, Kottkamp GS, Eppelbaum JL, Stubblefield BA, Gilbert SE, Gilbert ES. Influences of biofilm structure and antibiotic resistance mechanisms on indirect pathogenicity in a model polymicrobial biofilm. Appl Environ Microbiol 2006; 72: 5013-5019.
- Walsh C. Molecular mechanisms that confer antibacterial drug resistance. Nature 2000; 406: 775-781.
- Ong CL, Beatson SA, McEwan AG, Schembri MA. Conjugative plasmid transfer and adhesion dynamics in an Escherichia coli biofilm. Appl Environ Microbiol 2009; 75: 6783-6791.
- Varga M, Kuntova L, Pantucek R, Maslanova I, Ruzickova V, Doskar J. Efficient transfer of antibiotic resistance plasmids by transduction within methicillin-resistant Staphylococcus aureus USA300 clone. FEMS Microbiol Lett 2012; 332: 146-152.
- Krol JE, Nguyen HD, Rogers LM, Beyenal H, Krone SM, Top EM. Increased transfer of a multidrug resistance plasmid in Escherichia coli biofilms at the air-liquid interface. Appl Environ Microbiol 2011; 77: 5079-5088.
- Bjarnsholt T. The role of bacterial biofilms in chronic infections. APMIS Suppl 2013; : 1-51.
- Burmolle M, Thomsen TR, Fazli M, Dige I, Christensen L, Homoe P, Tvede M, Nyvad B, Tolker-Nielsen T, Givskov M, Moser C, Kirketerp-Moller K, Johansen HK, Hoiby N,Jensen PO, Sorensen SJ, Bjarnsholt T. Biofilms in chronic infections - a matter of opportunity - monospecies biofilms in multispecies infections. FEMS Immunol Med Microbiol 2010; 59: 324-336.
- Ghigo JM. Natural conjugative plasmids induce bacterial biofilm development. Nature 2001; 412: 442-445.
- Hennequin C, Aumeran C, Robin F, Traore O, Forestier C. Antibiotic resistance and plasmid transfer capacity in biofilm formed with a CTX-M-15-producing Klebsiella pneumoniae isolate. J Antimicrob Chemother 2012; 67: 2123-2130.
- Blumer C, Kleefeld A, Lehnen D, Heintz M, Dobrindt U, Nagy G, Michaelis K, Emody L, Polen T, Rachel R, Wendisch VF, Unden G. Regulation of type 1 fimbriae synthesis and biofilm formation by the transcriptional regulator LrhA of Escherichia coli. Microbiology, 2005; 151: 3287-3298.
- Ulett GC, Mabbett AN, Fung KC, Webb RI, Schembri MA. The role of F9 fimbriae of uropathogenic Escherichia coli in biofilm formation. Microbiology 2007; 153: 2321-2331.
- Bosse JT, Sinha S, Li MS, O'Dwyer CA, Nash JH, Rycroft AN, Kroll JS, Langford PR. Regulation of pga operon expression and biofilm formation in Actinobacillus pleuropneumoniae by sigmaE and H-NS. J Bacteriol 2010; 192: 2414-2423.
- Itoh Y, Rice JD, Goller C, Pannuri A, Taylor J, Meisner J, Beveridge TJ, Preston JF, Romeo T. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J Bacteriol 2008; 190: 3670-3680.
- Niu C, Robbins CM, Pittman KJ, Osborn L, Stubblefield BA, Simmons RB, Gilbert ES. LuxS influences Escherichia coli biofilm formation through autoinducer-2-dependent and autoinducer-2-independent modalities. FEMS Microbiol Ecol 2013; 83: 778-791.