Research Article - Current Pediatric Research (2017) Volume 21, Issue 4
Novel premature termination codon mutation in FBN1 gene in an infant with severe Marfan syndrome.
Brissia Lazalde1,2, Gerardo Aguirre-Hernández1, Rosa M González-Arreola1, José L Venegas Rodríguez1, Antonio E Gonzalez Font31Departamento de Genética, Facultad de Medicina y Nutrición, Universidad Juárez del Estado de Durango, Durango, Mexico.
2Unidad de Investigación Biomédica, Instituto Mexicano del Seguro Social, Durango, Mexico.
3Departamento de Cardiología Pediátrica, Hospital Materno-Infantil, Secretaría de Salud del Estado de Durango, Durango, México.
- Corresponding Author:
- Brissia Lazalde
Unidad de Investigación Biomédica
Instituto Mexicano del Seguro Social
Canoas 100, Col. Los Angeles, Durango
C.P. 34067, Mexico
Tel: +52-618-812-0997
E-mail: brissial@hotmail.com
Accepted date: August 30, 2017
Abstract
Marfan syndrome is a systemic disorder of connective tissue inherited in an autosomal dominant fashion due to mutations in the FBN1 gene. It is characterized by a broad clinical variability within and between families as well as in individuals. The clinical presentation in the early onset form of the disease is severe and rapidly progressive; deaths among infantile Marfan syndrome patients are associated with congestive heart failure due to severe mitral and/or tricuspid valvulopathy, although this severe presentation is rarely diagnosed and the majority of cases are sporadic. Here we report a case of an infant with severe cardiomyopathy diagnosed at 18 months of age. The clinical examination showed tall stature, dolichocephaly, long shaped facies, down slanting palpebral fissures, malar hypoplasia, high palate, protruding ears, pectus carinatum, scoliosis, articular hypermobility, long fingers and toes, hind foot deformity and myopia. A fibrillinopathy was suspected and molecular diagnostic tests of the TGFBR1, TGFBR2 and FBN1 genes were conducted. The direct analysis of the genes showed a heterozygous duplication of 4 nucleotides (c.5662_5665dupATGT) in FBN1 gene that creates a shift in the reading frame at codon 1889, producing a premature termination codon, 6 positions downstream (p.Cys1889Tyrfs*7). According to established criteria, it is a putative pathogenic mutation not reported previously. The mutation was not present in the parents. PTC mutations are commonly associated with milder phenotypes; therefore, this case highlights the complexity of the pathogenicity of FBN1 mutations. Molecular diagnosis in children suspected of having Marfan syndrome guarantees an early diagnosis, prognosis and timely therapy.
Keywords
FBN1, Marfan syndrome, Novel mutation, Premature termination codon mutation.
Background
Marfan syndrome (MFS; OMIM 154700) is a systemic disorder of connective tissue, with a prevalence of about 1/3300 to 1/5,000 individuals [1]. It is inherited in an autosomal dominant fashion and it is characterized by a broad phenotypic continuum ranging from neonatal presentation of severe and rapidly progressive disease in multiple organ systems to mild clinical entities [2]. The main features of this pleiotropic disorder include ectopia lentis (EL), skeletal manifestations such as disproportionate overgrowth of long bone and aortic root aneurysm [3].
The early onset and rapidly progressive form of MFS is atypical and rarely diagnosed; it is characterized by severe defects on the cardiovascular system, deaths among infantile MFS patients are associated with congestive heart failure due to mitral or tricuspid regurgitation [4].
Mutations in the central part of the FBN1 gene (NM 000138.4) between exons 24 and 32 have been associated with the most severe form of MFS that occurs in the first year of life [4-7]. Here, we report a case of an infant diagnosed with severe cardiomyopathy at 18 months of age in whom a novel premature termination codon (PTC) mutation in the FBN1 gene was detected. PTC mutations are commonly associated with milder phenotypes; therefore, this case highlights the complexity of the pathogenicity of FBN1 mutations and the importance of early detection of MFS in order to receive timely therapy.
Case Report
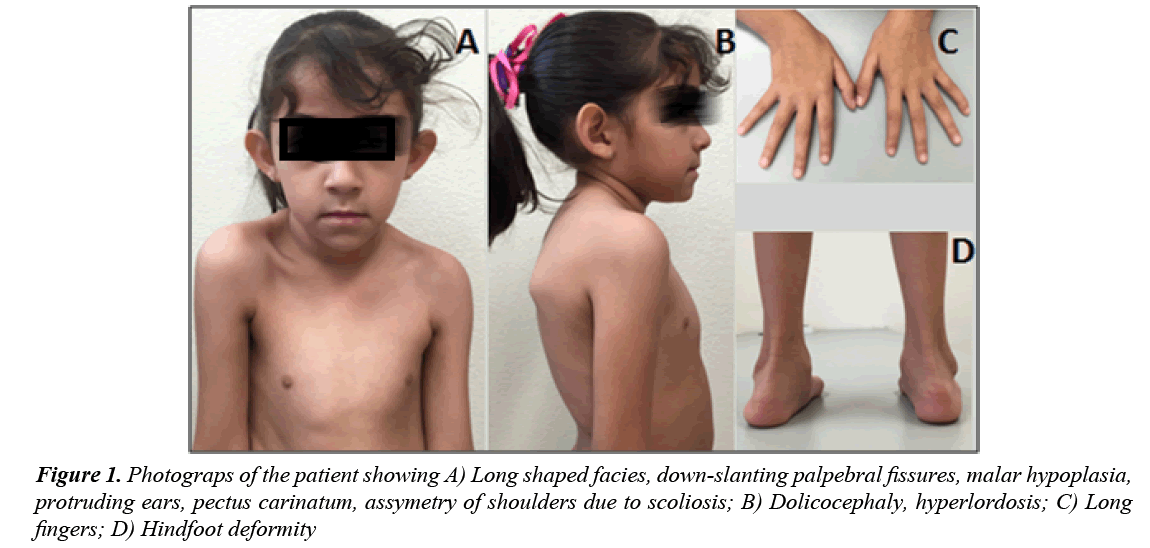
A 4 year old female infant was born to a 31 year old (gravida 4, parity 2) healthy mother and 33 year old healthy and unrelated father. She was born after uneventful pregnancy by vaginal delivery at 42 weeks of gestation; weight at birth was 3,250 g and length was 53 cm. At 18 months of age she was admitted to the emergency department due to respiratory infectious disease and the physical examination revealed a grade IV-VI holosystolic heart murmur. Therefore, an echocardiogram was performed revealing cardiomegaly, dilatation of left cavities and moderate to severe mitral insufficiency with mitral valve prolapse. Besides the echocardiography findings, dysmorphic features were noted and the patient was referred to the genetics service at the age of two years. The physical examination showed proportionate tall stature, height 94 cm (>97th percentile), weight 16.3 kg (25-50th percentile), dolichocephaly, long shaped facies, downslanting palpebral fissures, malar hypoplasia, high palate, protruding ears, pectus carinatum, scoliosis, articular hypermobility, long fingers of hand and feet and hind foot deformity (Figure 1). Ophthalmologic evaluation revealed myopia and excluded EL, but due to cardiac valvulopathy associated with marfanoid features, a fibrillinopathy was suspected. Genetic testing to identify pathogenic mutations of the FBN1, TGFBR1 and TGFBR2 genes were conducted in the patient after informed consent was obtained.
Genomic DNA was extracted from peripheral blood samples using Gentra Puregene blood kit (Qiagen, Hilden, Germany). The coding regions and splice junctions of the 3 genes were amplified using PCR. Libraries were prepared using the Nextera DNA Sample Prep Kit (Illumina). Sequencing was performed on the MiSeq Illumina platform. Bidirectional sequence was assembled and aligned to reference gene sequences based on human genome build GRCh37/UCSC hg19 and analyzed for sequence variants.
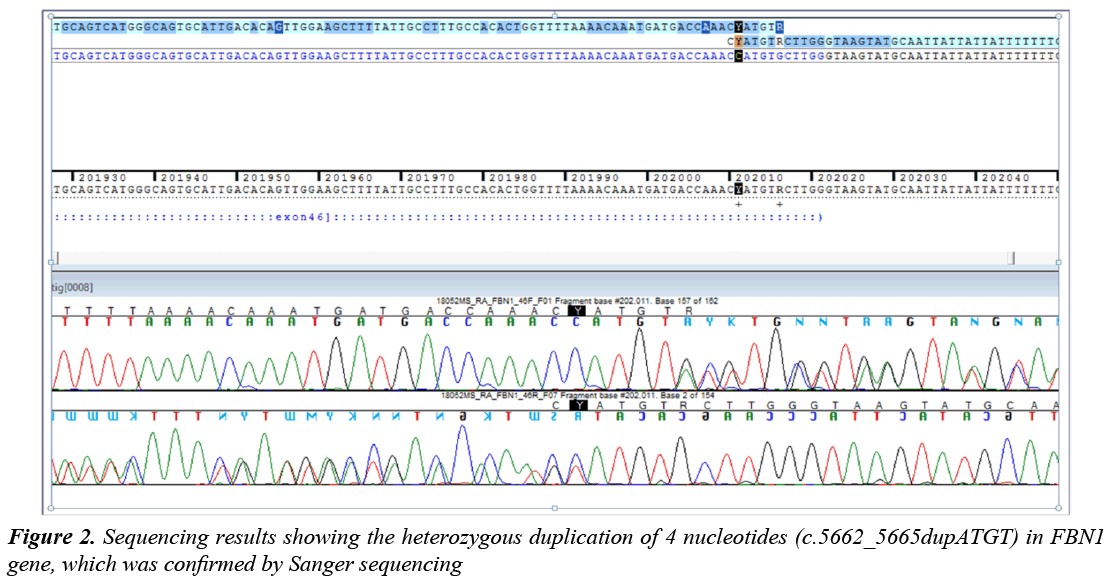
The direct analysis of the genes showed a heterozygous duplication of 4 nucleotides (c.5662_5665dupATGT) in exon 45 in FBN1 gene, which was confirmed by Sanger sequencing (Figure 2). Additionally, a heterozygous transition from G to A (C.6700G>A) that presumably produces, at protein level; a change of Valine at position 2234 by a Methionine (p.Val2234Met) has been detected in the same gene.
In order to find out if this mutation was de novo or inherited, parental and sibling genomic DNA was extracted from peripheral blood samples after informed consent was obtained. Direct Sanger sequencing of locus PCR products was conducted. The duplication c.5662_5665dupATGT was absent in the sibling and both parents.
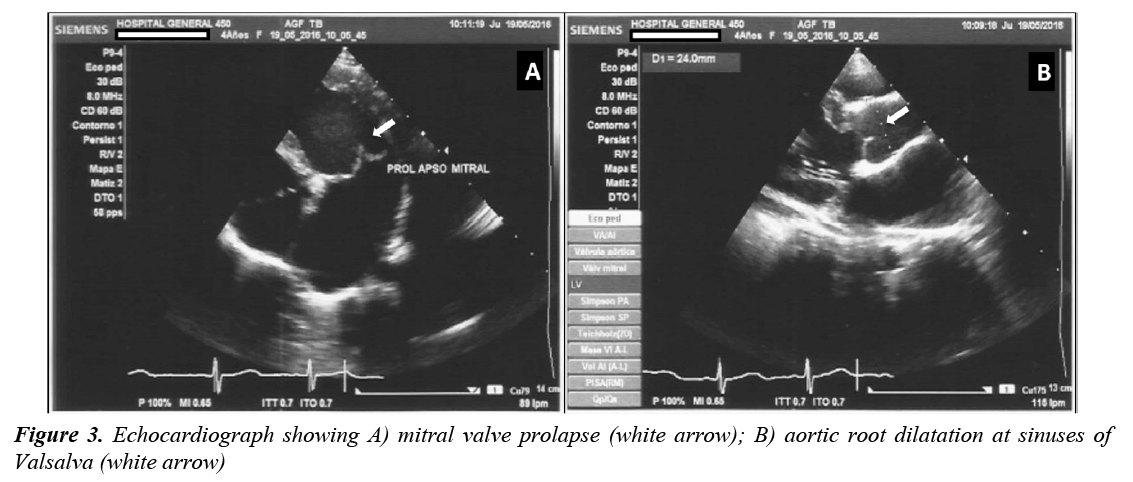
The last clinical examination, performed at the age of 4 years, showed an upper/lower segment ratio of 1.19 and arm span/height ratio of 1.04. The echocardiogram revealed dilated aortic root (24 mm, Z value ≥ +3), mitral and tricuspid valvular dysplasia, severe mitral insufficiency, redundant valves and tendinous chords with prolapse, showing mitral regurgitation jets that reached the roof of the left atrium, dilatation of left atrium and ventricle and diminished left ventricular longitudinal strain (Figure 3). Because a poor prognosis was established, the patient underwent mitral valve replacement performed using 27 mm St. Jude Medical mechanical valve with a favorable posoperative outcome.
Discussion
The patient was evaluated according to the revised Ghent criteria [3]. In the absence of any family history, as in the present case, aortic root aneurysm and ectopia lentis would be sufficient for the diagnosis of Marfan syndrome, but in the absence of these signs, the presence of an FBN1 mutation and positive systemic score is required.
The detected systemic features (scores) in the present case included: pectus carinatum deformity (2), hindfoot deformity (2), scoliosis or thoracolumbar kyphosis (1), 3 of 5 facial features (1), mitral valve prolapse (1) and severe myopia (1). These systemic features (score>8) combined with the presentation of severe aortic root dilation (Z>2) and the presence of a FBN1 mutation led us to the diagnosis of MFS.
To date 1847 different mutations in FBN1 have been documented on the Universal Marfan database-FBN1 (http://www.umd.be). There had been proposed different mechanisms of FBN1mutations pathogenesis. Mutations that result in haploinsufficiency produce cuantitative defects due to the reduction of the produced fibrillin, while dominant negative mutations lead to cualitative defects due to the change in the structure that may affect function such as folding or protein–protein interactions [8].
The mutation found in our patient has not been reported previously. According to the in silico analysis, the duplication of 4 nucleotides (c.5662_5665dupATGT) creates a shift in the reading frame at codon 1889, producing a premature termination codon (PTC) 6 positions downstream (p.Cys1889Tyrfs*7). The pathogenic nature of this mutation was assessed using established criteria [3,9].
PTC mutations commonly generate truncated transcripts that result in degradation by the nonsense-mediated mRNA decay pathway with mutant transcript levels ranging from 2%-28% compared with the normal transcript [10]. In a study comparing protein phenotypes in fibroblast cultures it was observed that probands with PTC mutations have markedly reduced extracellular fibrillin deposition with reduced synthesis [10]. These mutations are underrepresented in patients with severe phenotypes; according to a study in children with MFS, PTC mutations were only present in 25% of cases compared to 36% in adults [11]. In the present case the phenotype of the patient was severe, since she was diagnosed early in infancy with severe cardiovascular manifestations.
The most severe forms of MFS are commonly due to mutations clustered in the center portion of the gene (exons 24-32), so-called “neonatal region”, with overrepresentation of missense mutations [9]. Only 6% of the patients with neonatal or severe MFS have mutations in other regions of the FBN1 gene [9]. Despite the severe presentation of the present case, the mutation was found out of the “neonatal region” in exon 45 of FBN1 gene, in the calcium binding consensus sequence 27 (cb EGF-like #27). According to UMD-FBN1 mutation database, there are only two reports of PTC mutations located in the exon 45; one of them had major effect on the cardiac system and the second is associated with a classical MFS [12,13]. Therefore it is evident that besides the location of the stop codon, there must be other modifiers that contribute to the severity of the phenotype such as allelic variation in the normal FBN1 gene, mild mutations in FBN1, TGFBR1 and TGFBR2 allele, mutations in other organ specific genes, as well as epigenetic mechanisms and possibly environmental factors [14-18].
In addition to the pathogenic mutation, a rare polymorphism was found (C.6700G>A, rs112084407) with low frequency of heterozygotes and uncertain clinical significance, although the in silico analysis with SIFT, Mutation Taster and software Polyphen predict that change is probably benign.
It has been proposed that expression of wild type allele below a critical threshold could result in a more profound disease [12,14]. We did not conduct qualitative analysis of FBN1 mRNA, therefore we cannot speculate if low expression levels of the wild type allele may have contributed to the severity of the disease.
The mutation in the present case is de novo because it is not present in the apparently healthy parents, which is common in the early onset presentation of the disease, since patients with de novo mutations tend to be more seriously affected and may not survive to reproduce [11].
In cases of children with sporadically occurring disease, the MFS diagnosis could be difficult since many manifestations such as EL which is a major criterion, are age dependent. Aditionally, it is important to consider that although there is no strict genotype-phenotype correlation, the clinical manifestations could depend on the type of mutation; therefore FBN1 mutation testing in children suspected of having MFS or other related type 1 fibrillinopathy guarantees an early diagnosis, prognosis and timely therapy.
Acknowledgement
The authors are grateful to the mother of the patient, who provided written informed consent for the case publication, Thank you to Dr. Ramiro Ramirez-Solis for his review of the manuscript.
References
- Arslan-Kirchner M, Arbustini E, Boileau C, et al. Clinical utility gene card for: Marfan syndrome type 1 and related phenotypes (FBN1). Eur J Hum Genet 2010; 18: 1-15.
- https://www.ncbi.nlm.nih.gov/books/NBK1335/
- Loeys BL, Dietz HC, Braverman AC, et al. The revised Ghent nosology for the Marfan syndrome. J Med Genet 2010; 47: 476-485.
- Tiecke F, Katzke S, Booms P, et al. Classic, atypically severe and neonatal Marfan syndrome: twelve mutations and genotype-phenotype correlations in FBN1 exons 24-40. Eur J Hum Genet 2001; 9:13-21.
- Ramirez F, Dietz HC. Marfan syndrome: from molecular pathogenesis to clinical treatment. Curr Opin Genet Dev 2007; 17: 252-258.
- Booms P, Cisler J, Mathews KR, et al. Novel exon skipping mutation in the fibrillin-1 gene: two 'hot spots' for the neonatal Marfan syndrome. Clin Genet 1999; 55: 110-117.
- Peng Q, Deng Y, Yang Y, Liu H. A novel fibrillin-1 gene missense mutation associated with neonatal Marfan syndrome: a case report and review of the mutation spectrum. BMC Pediatrics 2016; 16: 60.
- Landis BJ, Veldtman GR, Ware SM. Genotype–phenotype correlations in Marfan syndrome. Heart 2017.
- Faivre L, Collod-Beroud G, Loeys BL, et al. Effect of mutation type and location on clinical outcome in 1,013 probands with Marfan syndrome or related phenotypes and FBN1 mutations: an international study. Am J Hum Genet 2007; 81: 454-466.
- Schrijver I, Liu W, Odom R, et al. Premature termination mutations in FBN1: distinct effects on differential allelic expression and on protein and clinical phenotypes. Am J Hum Genet 2002; 71: 223-237.
- Faivre L, Masurel-Paulet A, Collod-Béroud G, et al. Clinical and molecular study of 320 children with Marfan syndrome and related type I fibrillinopathies in a series of 1009 probands with pathogenic FBN1 mutations. Pediatrics 2009; 123: 391-398.
- Tjeldhorn L, Amundsen SS, Barøy T, et al. Qualitative and quantitative analysis of FBN1 mRNA from 16 patients with Marfan Syndrome. BMC Med Genet 2015; 16: 113.
- Biggin A, Holman K, Brett M, et al. Detection of thirty novel FBN1 mutations in patients with Marfan syndrome or a related fibrillinopathy. Hum Mutat 2004; 23: 99.
- Hutchinson S, Furger A, Halliday D, et al. Allelic variation in normal human FBN1 expression in a family with Marfan syndrome: a potential modifier of phenotype? Hum Mol Genet 2003; 12: 2269-2276.
- Lucarini L, Sticchi E, Sofi F, et al. ACE and TGFBR1 genes interact in influencing the susceptibility to abdominal aortic aneurysm. Atherosclerosis 2009; 202: 205-210.
- Lucarini L, Evangelisti L, Attanasio M, et al. May TGFBR1 act also as low penetrance allele in Marfan syndrome? Int J Cardiol 2009; 131: 281-284.
- Gomez D, Coyet A, Ollivier V, et al. Epigenetic control of vascular smooth muscle cells in Marfan and non-Marfan thoracic aortic aneurysms. Cardiovascular Research 2011; 89: 446-456.
- Robinson PN, Godfrey M. The molecular genetics of Marfan syndrome and related microfibrillopathies. J Med Genet 2000; 37: 9-25.