Research Article - Biomedical Research (2018) Materials and Image Processing Techniques for Biomedical Applications
Non-rigid registration of medical images using a hierarchical framework with normalized mutual information
Min Li1*, Limei Zhang1, Nan Hu2 and Zhikang Xiang1
1School of Computer Science and Engineering, Nanjing University of Science and Technology, Nanjing, PR China
2Department of Radiology, Daping Hospital of Third Military Medical University, Chongqing, PR China
- *Corresponding Author:
- Min Li
School of Computer Science and Engineering
Nanjing University of Science and Technology
PR China
Accepted date: October 5, 2017
DOI: 10.4066/biomedicalresearch.29-17-1995
Visit for more related articles at Biomedical ResearchAbstract
Medical image registration plays a crucial role in medical field. Existing medical registration methods can be categorized as rigid registration and non-rigid registration methods. Compared with rigid registration, the non-rigid registration is more suitable for representing the anatomy’s complicated deformation. In this study, we develop a non-rigid registration method in which Normalized Mutual Information (NMI) is used as similarity measurement and B-splines are used for transformation. The algorithm is implemented using a hierarchical framework such that the accuracy of registration is improved by the coarse-to-fine registration schema. Experiments demonstrate that the proposed method produces more accurate results than the affine and demons methods in registration of pelvis CBCT images, which provides an alternative approach to support non-rigid registration of medical images with intensity variation. The proposed algorithm shows potential for facilitating image-guided radiation therapy with higher treatment dose delivered to a precise location through the use of image registration.
Keywords
Normalized mutual information (NMI), Non-rigid registration, Hierarchical framework
Introduction
Medical image registration is a very important research topic in the field of image processing, which can be taken as a technical basis for many other research such as medical image fusion and medical image reconstruction because medical image registration aims to establish the spatial correspondence between images acquired at different time. Existing registration methods can be divided into rigid registration and non-rigid registration approaches [1]. Rigid methods are suitable for alignment of the rigid object such as bony anatomy and generally present in commercial medical image analysis packages [2]. Some tools have been developed and applied for rigid registration, such as NiftyReg [3] and FLIRT [4].
In medical education and clinical practice, registration of pelvis images is of great significance. For instance, during pelvic radiotherapy, pelvis image registration plays a crucial role in locating tumors and calculating dose to deliver. However, pelvis image registration suffers from inter-fraction changes, such as variation of patients’ positions and status of bladder filling between image acquisitions at different time, which cause registration of pelvis images rather difficult. Moreover, pelvic organs may be deformed due to patients’ respiratory motion. As such, simple rigid methods are not suitable to use while non-rigid methods have more potentials in handling with registration of the anatomy with complex deformation.
In the past decades, a large number of methods have been proposed for non-rigid registration, such as feature-based algorithms [5-7], intensity-based approaches [8-10], and physical models [11,12]. However, very few methods focus on registration of pelvis images with reasonable results. For instance, Gao et al. [13] introduced an appearance embedding statistical deformation model which was built using the node positions of deformations obtained from registration of pelvis CT images and applied for bony pelvis segmentation from MRI images. To match the femur and pelvis from 3D ultrasound images, Barratt et al. [14] matched bones by manually adjusting parameters that defined a rigid-body transformation with scaling, which provided initial estimates for the followed intensity-based non-rigid registration. It should be noted that the surface of the bone of interest is delineated semi-automatically before registration which is a time-consuming process and the registration accuracy also relies on the segmentation quality. At present, registration of pelvis images is still a problem to solve in medical education and clinical practice, and it is necessary to improve the accuracy of registration for particular clinical applications, which provides a strong motivation for developing new technology to improve registration efficacy and accuracy.
In this study, we present a hierarchical framework in which we use Normalized Mutual Information (NMI) as the similarity measure and B-splines as transformation for non-rigid registration of clinical images. The proposed method makes full use of the advantages of the multi-resolution schema and obtains reasonable registration results through a coarse-to-fine process.
Methods
Among non-rigid registration, intensity based algorithms such as demons and optical flow methods have been widely applied. However, both demons and optical flow methods are used under assumption that the image intensity keeps conserved from one moment to another. Differently, the MI is not limited to such an assumption of intensity conservation and able to use as a similarity measurement handling with intensity variation.
Given two images A and B, the MI of two images expresses how much the uncertainty on one of the image decreases when the other one is known. Therefore, the MI is assumed to be maximum when the two images are well matched. However, the poor registration may cause a high MI value which has been found in previous publication [15]. Therefore, we use NMI as the similarity metric which is defined as:
NMI (A, B)=H (A)+H (B)/H (A, B) → (1)
where H means the Shannon entropy and H (A, B) is the Shannon entropy of the two images joint distribution.
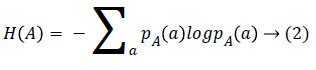
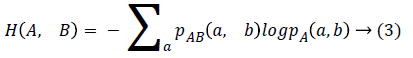
We aims to align A and B images by minimizing the Equation 1. Actually, image registration can be described as a problem of finding the displacement vectors for the Region of Interest (ROI). This allows us to use cubic B-splines as the transformation such that the displacement vector of a voxel in image can be controlled by a small number of control points. The displacement of a voxel I can be defined using cubic Bsplines as [16]:

where ϕj are parameters which represent the displacements of control points and are B-spline basis functions.
We divide the registration process into several registration layers which form a hierarchical framework. Specifically, we initially sample the original image with a uniform space interval n such that the original image is resized with a coarse grid. We then apply the NMI based registration method in the first layer. The resulting deformation field is then used as initial condition for the registration of the next layer of the hierarchy. In the next layer, images are sampled with space interval n/2 which leads to a less coarse image grid for registration. The above process is repeated with the space interval reduced as the half value of the previous layer until the predefined layer number reached. In each layer, the NMI based registration is same and implemented with the registration result used in the next layer.
Results and Discussion
From March 2013 to August 2014, 55 female patients with cervical cancer received radiotherapy and their CBCT images were acquired at Daping Hospital of Third Military Medical University in China. The patients were placed in the supine position on a flat couch and CBCT images were obtained before treatment. Each patient was imaged more than 10 times in different days and the 3D CBCT data was reconstructed every time. Each 3D CBCT image is composed of 40-60 slices and reconstructed on 512 × 512 matrix. Each image voxel represents a special resolution of 0.6 mm × 0.6 mm × 2.5 mm. We randomly selected ten pairs of CBCT images to conduct our registration experiment and applied the proposed method to match pelvis CBCT images in this study. The scanner table was removed according to its position in each image prior to registration.
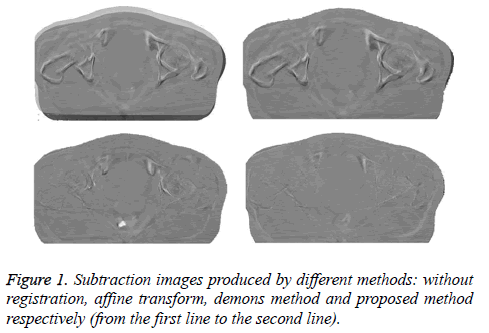
To evaluate the registration performance, we create subtraction images by subtracting the deformed reference image from the target image. Figure 1 shows the subtraction images created by different registration methods including affine transformation, demons method and the proposed algorithm. Difference has been enhanced in the direct subtraction image before registration and reduced after registration. Compared with the subtraction image produced by affine transformation and demons method, the proposed algorithm achieves much better subtraction image with only very minor mismatch, which indicates that the proposed algorithm is promising for registration of pelvis CBCT images.
We calculate the width of the gray-scale histogram to evaluate the quality of the subtraction images produced by different methods (as shown in Table 1). In general, the narrower the histogram width, the better the quality of the subtraction image [17]. The subtraction images have a large Mean Histogram Width (MHW) of 144.56 before registration, which is reduced to 115.56, 86.27 and 69.89 respectively for affine transformation, demons method and the proposed algorithm. In addition, by calculating the NMI, we obtain the mean NMI (MNMI) of 0.66 before registration which is improved to 0.75 and 0.89 respectively for affine and demons methods. However, the proposed method achieves the highest MNMI of 0.93, which is more suitable than the other two methods for registration of pelvis CBCT images.
| Measurement | Registration results using different methods | |||
|---|---|---|---|---|
| No registration | Affine | Demons | Proposed | |
| MHW | 144.56 | 115.56 | 86.27 | 69.89 |
| MNMI | 0.66 | 0.75 | 0.89 | 0.93 |
Table 1. Mean Histogram Width (MHW) of subtraction images and mean Normalized Mutual Information (MNMI) produced by different methods for registration of pelvis CBCT images.
Conclusion
In this paper, we presented a non-rigid registration method using a hierarchical framework in which the normalized mutual information is used as similarity measure and B-splines are used for transformation. Differently from many methods limited to intensity conservation, the proposed method is able to handle registration of images with intensity variation. Experiments show that the proposed method archives more accurate and reasonable registration results than affine and demons methods in alignment of pelvis CBCT images, providing an alternative way to support non-rigid registration of medical images. Moreover, the proposed algorithm shows potentials for facilitating image-guided radiation therapy in which higher treatment dose can be delivered to a precise location through the use of image registration. Our future work will focus on extension of the proposed algorithm and its application to multimodal image registration.
Conflict of Interest
The authors declare that there are no competing interests regarding the publication of this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 61501241), the Natural Science Foundation of Jiangsu Province (No. BK20 150792), and the project funded by China Postdoctoral Science Foundation (No. 2015 M570450).
References
- Muyan-Ozcelik P, Owens JD, Xia J, Samant SS. Fast deformable registration on the gpu: a cuda implementation of demons. Proc Int Conf Comp Sci Appl 2008.
- Viergever MA, Maintz JBA, Klein S. A survey of medical image registration-under review. Med Imag Anal 2016; 33: 140-144.
- Ourselin S, Roche A, Subsol G, Pennec X, Ayache N. Reconstructing a 3D structure from serial histological sections. Image Vis Comp 2001; 19: 25-31.
- Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. Neuroimage 2012; 62: 782-790.
- Lobachev O, Ulrich C, Steiniger BS. Feature-based multi-resolution registration of immunostained serial sections. Med Image Anal 2017; 35: 288-302.
- Verhoeven T, Tong X, Schreurs R. Quantification of facial asymmetry: a comparative study of landmark-based and surface-based registrations. J Cranio-Maxillofac Surg 2016; 44: 1131-1136.
- Marina S, Papp EA, Rembrandt B. Anatomical landmarks for registration of experimental image data to volumetric rodent brain at lasing templates. J Neurosci Methods 2015; 240: 161-169.
- Ghafurian S, Hacihaliloglu I, Metaxas DN. A computationally efficient 3D/2D registration method based on image gradient direction probability density function, Neurocomp 2017; 229: 100-108.
- Valsecchi A, Damas S, Santamaria J. Intensity-based image registration using scatter search. Artificial Intel Med 2014; 60: 151-163.
- Aghajani K, Manzuri MT, Yousefpour R. A robust image registration method based on total variation regularization under complex illumination changes. Com Methods Progs Biomed 2016; 134: 89-107.
- Pawar A, Zhang YJ, Jia Y. Adaptive FEM-based nonrigid image registration using truncated hierarchical B-splines. Comp Math Appl 2016; 72: 2028-2040.
- Hopp T, Duric N, Ruiter NV. Image fusion of ultrasound computer tomography volumes with X-ray mammograms using a biomechanical model based 2D/3D registration. Comp Med Imag Graphs 2015; 40: 170-181.
- Gao Q, Chang PL, Rueckert D. Modeling of the bony pelvis from MRI using a multi-atlas AE-SDM for registration and tracking in image-guided robotic prostatectomy. Comp Med Imag Graph 2013; 37: 183-194.
- Barratt DC, Chan CSK, Edwards PJ. Instantiation and registration of statistical shape models of the femur and pelvis using 3D ultrasound imaging. Med Imag Anal 2008; 12: 358-374.
- Gao Z, Gu B, Lin J. Monomodal image registration using mutual information based methods. Image Vis Comp 2008; 26: 164-173.
- Szeliski R, Coughlan J. Spline-based image registration. Int J Comp Vis 1997; 22: 199-218.
- Armato SG, Doshi DJ, Engelmann R. Temporal subtraction in chest radiography: automated assessment of registration accuracy. Med Phys 2006; 33: 1239-1249.