Research Article - International Journal of Pure and Applied Zoology (2017) Volume 5, Issue 3
MOLECULAR TYPING OF WATERBORNE E. COLI STRAINS USING REP-PCR
- *Corresponding Author:
- Maryam Makhmalzadeh
Department of Microbiology, Faculty of Science
Karaj Branch, Islamic Azad University, Karaj, Iran
E-mail: maryam_makhmalzadeh@yahoo.com
Received 01st June 2017; Accepted 17th July 2017; Published 21st July 2017
Abstract
Water borne bacteria, including E. coli as the top bacterial quality indicator, are over to be the vital concerns of some society. Typing of bacterial strains is used to confirm or reject epidemiologic evidence that a particular bacterium is the source of infection in a food type, water source or nosocomial infection. Molecular typing is a valued instrument for analysis of genetic relationship among the microbial strains. The purpose of the present study was to determine the molecular types of waterborne E. coli strains using REP-PCR. Present study comprised 100 waterborne E. coli strains isolated from different water sources in Karaj, Iran in 2013. Bacterial isolates were detected and identified via standard microbiological and biochemical exams. Genomic DNA was extracted via Genomic DNA Extraction Kit and genetic relationship among the strains was evaluated via REP-PCR using REP1 and REP2 primers. PCR amplicons were electrophoresed in 1.5% agarose gel and staining using ethidium bromide and visualized via a Gel DocTM XR+(BIORAD) and dendrogram was constructed based on Dice Comparison method and UPGMA Clustering. Using REP-PCR, all strains were typeable. Over 15 different bands ranging from 130 to 2300 bp were amplified in different profiles. Following digitizing (0 and 1) the positive genes and dendrogram construction by PAUP 4.0 software, dendrogram analysis showed the REP-PCR differentiated 100 environmental E. coli strains into 8 rep clusters however the r6 (28%) was the most prevalent cluster. Results of the present study indicated that E. coli strains isolated from different water sources in Karaj belong to diverse clones and different genotypes. Our finding also showed that REP-PCR is a powerful molecular tool with high performance and good discriminatory power for molecular typing of waterborne E. coli.
Keywords
E. coli; Molecular typing; Repetitive sequence-based PCR
Abbreviations
E. coli: Escherichia coli; UTIs: Urinary Tract Infections; ETEC: Enterotoxigenic; EHEC: Enterohemorrhagic; EIEC: Enteroinvasive.
Introduction
Waterborne diseases have a negative impact on public health in developing countries, where several people do not have access to a safe drinking water source and, therefore, several die of waterborne bacterial infections (Cabral, 2010; Ranjbar et al., 2016; Ranjbar et al., 2017a). The presence of Escherichia coli (E. coli) in drinking water is a significant concern for public health (Hunter, 2003; Tajbakhsh et al., 2015; Ranjbar et al., 2017 b). Escherichia coli is a commensal member of the intestinal flora of humans and many animal hosts. Several genotypes have acquired specific virulence factors and are capable of causing disease as gastrointestinal diseases, urinary tract infections (UTIs) and sepsis/meningitis (Nataro and Kaper, 1998; Kaper et al., 2004; Anvarinejad et al., 2012; Tajbakhsh et al., 2016). E. coli strains isolated from intestinal diseases have been grouped into at least 6 different chief groups, based on epidemiological sign, clinical features, phenotypic characters of the disease and specific virulence factors. Enterotoxigenic enterohemorrhagic (EHEC, namely O157), (ETEC, namely O148) and enteroinvasive serotypes (EIEC, namely O124) are of outstanding significance and can be transmitted over polluted water (Bettelheim, 2003; Scheutz and Strockbine, 2005; Torkan et al., 2016).
It is a well-known fact that E. coli happens primarily in the gastrointestinal tract of animals and humans. Therefore, it is also present in natural environment especially in water, soil, and on plants. The spread of E. coli in the environment is also affected by discharge of municipal sewage into surface water and soil (Watkinson et al., 2007; Hemmatinezhad et al., 2015; Jahandeh et al., 2015; Kheiri et al., 2016). The identification of E. coli in water is an implicit indicator of fresh fecal contamination and consequently of the hazard of cooccurrence of enteric pathogens that can cause infection in susceptible populaces (Yates, 2007). Several rules estimate and mandate agreement by recreational and drinking water quality standards on the basis of the large quantity and incidence of E. coli (Dufour, 1984; NRCC, 2004).
The spreading and fate of a species in a regular environment may possibly, in measure, be governed via range in the species; therefore, approximating this range is necessary. Some highresolution molecular fingerprinting methods have been used to make public species and subspecies variety (Rademaker et al., 2000; Schloter et al., 2000; Vaneechoutte, 1996). As many as 291 and 94 rep-PCR genotypes were well-known in collections of 643 river isolates and 353 beach water E. coli isolates, respectively (McLellan, 2004). The incidence of pathogenic E. coli in environmental water creates a potential risk for infections in humans and animals especially since water is used for irrigation, a source of drinking water, and for recreational purposes (Hamelin et al., 2007; Kümmerer, 2009; Koczura et al., 2012). At this time, molecular biologybased techniques are used for epidemiological examinations for the control and of outbreaks and monitoring of the spread of potential pathogens (Rahimi et al., 2012; Raissy et al., 2014; Rahimi et al., 2017).
However, despite the high incidence of waterborne diseases in this country, there are no published data on the determination of molecular types of waterborne E. coli strains using REP-PCR. Therefore, the purpose of this study was to determine the presence of waterborne E. coli strains and describe the strain diversity of an E. coli population retrieved from different water sources in Karaj by using a REP-PCR method. Therefore, our results will help to determine E. coli strains in water and to estimate the significance of these organisms for public health. Information obtained in this study will help to determine whether water of the examined reservoir contains a variety of E. coli genotypes and diversity strains of E. coli populations that can pose epidemiological threat.
Materials and Methods
Present study contained within 100 waterborne E. coli strains isolated from different water sources in Karaj, Iran in 2013. Bacterial isolates were detected and identified by standard microbiological and biochemical tests. Genomic DNA was extracted through AccuPrep® Genomic DNA Extraction Kit.
REP-PCR conditions and primers
REP-PCR primer sequences: Rep1, 5'-IIIICGICATCIGGC- 3' and Rep 2. 5'-ICGICTTATCIGGCCTA-3' and the PCR reaction conditions were as described through Versalovic et al. (1991), in a final volume of 50 ml, by minor modifications as follows: an initial denaturation (94ºC, 7 min) followed via 30 cycles of denaturation (90ºC, 30 s), annealing (40ºC, 1 min), and extension (72ºC, 8 min) by a single final extension (72ºC, 15 min). The size of the amplified fragments was visualized through electrophoresis in submersed agarose gel (1.5%) by 100 bp and 1 kb DNA markers (Life Technologies) as standards. The PCR for each strain was performed in three separate experiments to confirm the pattern of amplified bands.
Agarose gel electrophoresis
Agarose (1.5%) gel electrophoresis was done as described via (Sambrook et al., 1989).
Fingerprint analyses
Rep-PCR DNA fingerprinting of amplified DNA fragments obtained through agarose gel electrophoresis were verified. The incidence of a given band was coded as 1 and the lack of a given band was coded as 0 in a data matrix and analyzed using the PAUP 4.0 software. Dendrograms of dissimilarity were constructed for separate case.
Results
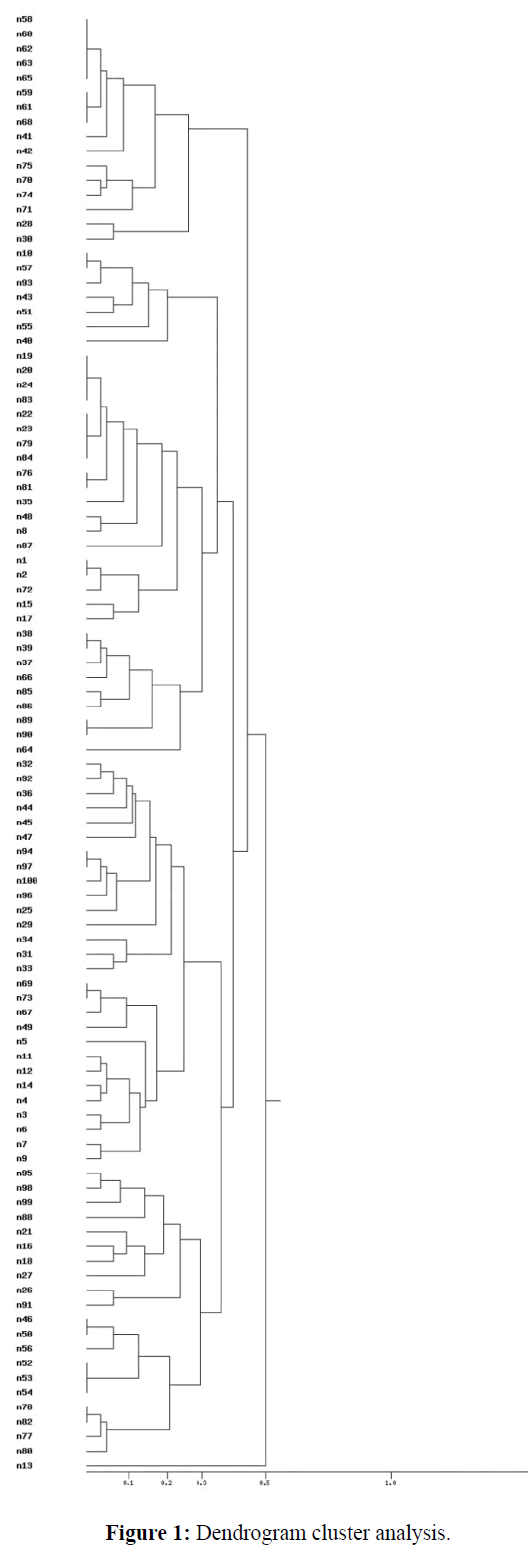
By Rep-PCR, all strains involved in this study were typeable. Over 15 different bands ranging from 130 to 2300 bp examined in this study were amplified in different profiles. Overall of 100 E. coli isolates were included in this study, of which majority of the isolates (28%) were categorized within 6 Rep clusters (Table 1). Following dendogram analysis (Figure 1), Rep-PCR could categorize the strains within 9 Rep clusters (Table 1).
| Cluster | 1r | 2r | 3r | 4r | 5r | 6r | 7r | 8r |
| Number | 14 | 2 | 7 | 19 | 9 | 28 | 10 | 11 |
| Percent | 14% | 2% | 7% | 19% | 9% | 28% | 10% | 11% |
Table 1: Percentages of E. coli isolates.
Dendrogram (Figure 1), which showed the results of cluster analyses aiming at determining most closely related isolates, as well as verification of their potential relationship with the reference strain, were constructed based on the rep- PCR analyses. The figures show less significant diversity between E. coli isolates. Based on these dendrograms, it can be concluded that the examined isolates are characterized by small genetic diversity and they can be easily grouped into clusters.
Discussion
Ensuring water safety is an ongoing challenge to public health providers. Evaluating the presence of fecal contamination indicators in water is important to defend public health from diseases caused by waterborne pathogens (Mendes Silva and Domingues, 2015). In the current study, overall of 100 E. coli isolates were recovered from different water sources. Overall, a relative high genotypic diversity was observed among the waterborne E. coli isolates. Small water systems that supply rural townships or camps have commonly been associated with waterborne outbreaks (Olsen et al., 2002).
The result of water borne strains with the similar genotypes, and the relationship of virulence gene profiles among strains, recommends that transmission of E. coli may happen among animals and humans or both host species are infected via a public source. Naturalized E. coli populaces have been identified in a range of environments, such as water or soil, and in tropical, temperate, or cold regions (Byappanahalli et al., 2006; Beversdorf et al., 2007; Byappanahalli et al., 2007; Ishii et al., 2007).
Escherichia coli genotypic and phenotypic diversity is believed to be extended (Ishii and Sadowsky, 2008), and it has been recommended that collections of as various as 40,000 isolates might be essential in order to capture all of the E. Coli diversity based on rep-PCR DNA fingerprinting (Johnson et al., 2004). The result of our study showed, Rep-PCR could categorize the strains within 9 Rep clusters. Several characteristics of this enormous diversity have significance for the valuation and organization of water quality. Rep-PCR is a microbial source tracking (MST) technique generally employed to elucidate the source of fecal contamination of surface water (Johnson et al., 2004; EPA, 2005; Edge and Schaefer, 2006).
REP-PCR was a number of as the molecular typing method for the E. coli isolates since it is rapid, reproducible, easy to perform, and highly discriminatory at the subspecies level (Olive and Bean, 1999), yielding results that compare well by pair wise DNA-DNA analyses (Rademaker et al., 2000). The present study presented, clusters were famous via host species nonetheless did depend powerfully on virulence gene profiles. Earlier studies that have used microbial source tracking (MST), PFGE, and whole genome sequencing to compare O26 strains from food animals and human have establish like results (Leomil et al., 2005; Ju et al., 2012).
In additional lake study showed on 11 sites over a 9-month period, repPCR genotyping of E. coli isolates showed that a little E. coli genotypes consistently dominated populations recovered from the area (Walk et al., 2007). As it has been described before, phages infecting distantly related bacterial hosts usually share slight or no nucleotide sequence similarity, even though phages infecting a specific bacterial host are additional similar (Hatfull, 2008).
Escherichia coli isolates examined in this study are characterized by small genetic diversity and they can be easily grouped into clusters. REP-PCR is a powerful molecular tool with high performance and good discriminatory power for molecular typing of waterborne E. coli. Therefore, the results show that it is urgent to evaluate the management of different water sources and their quality in Karaj to prevent the emergence of infectious outbreaks.
Competing Interests
The authors declare that they have no competing interests.
Acknowledgement
We would like to thank from the “Clinical Research Development Center of Baqiyatallah hospital” for their kindly cooperation.
References
- Anvarinejad, M., Farshad, S. H., Ranjbar, R., Giammanco, G. M., Alborzi, A. and Japoni A., 2012. Genotypic Analysis of E. coli Strains Isolated from Patients with Cystitis and Pyelonephritis. Iran. Red. Crescent. Med. J. 14: 408-416.
- Bettelheim, K. A., 2003. The genus Escherichia. In The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community, 3th ed.; Dworkin, M., Falkow, S., Rosenberg, E., Eds.; Springer-Verlag: New York, NY, USA.
- Beversdorf, L. J., Bornstein-Forst, S. M. and McLellan, S. L., 2007. The potential for beach sand to serve as a reservoir for Escherichia coli and the physical influences on cell die-off. J. Appl. Microbiol. 102: 1372-1381.
- Byappanahalli, M. N., Whitman, R. L., Shively, D. A., Ferguson, J., Ishii, S. and Sadowsky, M. J., 2007. Population structure of Cladophora-borne Escherichia coli in near shore water of Lake Michigan. Water. Res. 41: 3649-3654.
- Byappanahalli, M. N., Whitman, R. L., Shively, D. A., Sadowsky, M. J. and Ishii, S., 2006. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ. Microbiol. 8: 504-513.
- Cabral, J. P. S., 2010. Water microbiology. Bacterial pathogens and water. Int J Environ Res Public Health. 7: 3657-3703.
- Dufour, A. P., 1984. Health effects criteria for fresh recreational waters. Office of Research and Development, U.S. Environmental Protection Agency, Research Triangle Park, NC.
- Edge, T. A. and Schaefer, K. A., 2006. Microbial source tracking in aquatic ecosystems: the state of the science and an assessment of needs. National Water Research Institute, Burlington, Ontario, Canada.
- Environmental Protection Agency, 2005. Microbial source tracking guide document. EPA/600/R-05/064. National Risk Management Research Laboratory, Office of Research and Development, U.S. EPA, Cincinnati, OH.
- Hamelin, K., Bruant, G., El-Shaarawi, A., Hill, S., Edge, T. A., Fairbrother, J., Harel, J., Maynard, Cet., Masson, L. and Brousseau, R., 2007. Occurrence of virulence and antimicrobial resistance genes in Escherichia coli isolates from different aquatic ecosystems within the St. Clair River and Detroit River areas. Appl. Environ. Microbiol. 73: 477-484.
- Hatfull, G.F., 2008. Bacteriophage genomics. Curr. Opin. Microbiol. 11: 447-453.
- Hemmatinezhad, B., Khamesipour, F., Mohammadi, M., Safarpoor Dehkordi, F. and Mashak, Z., 2015. Microbiological Investigation of O-Serogroups, Virulence Factors and Antimicrobial Resistance Properties of Shiga Toxin-Producing Escherichia Coli Isolated from Ostrich, Turkey and Quail Meats. J Food Saf. 35: 491-500.
- Hunter, P., 2003. Drinking water and diarrheal disease due to Escherichia coli. J. Water Health 1: 65-72.
- Ishii, S. and Sadowsky, M. J., 2008. Escherichia coli in the environment: implications for water quality and human health. Microbes Environ. 23, 101-108.
- Ishii, S., Hansen, D. L., Hicks, R. E. and Sadowsky, M. J., 2007. Beach sand and sediments are temporal sinks and sources of Escherichia coli in Lake Superior. Environ. Sci. Technol. 41: 2203-2209.
- Jahandeh, N., Ranjbar, R., Behzadi, P. and Behzadi, E., 2015. Uropathogenic Escherichia coli virulence genes: invaluable approaches for designing DNA microarray probes. Cent. European J. Urol 68: 452-458.
- Johnson, L. K., Brown, M. B., Carruthers, E. A., Ferguson, J. A., Dombek, P. E. and Sadowsky, M. J., 2004. Sample size, library composition, and genotypic diversity among natural populations of Escherichia coli from different animal’s influence accuracy of determining sources of fecal pollution. Appl. Environ. Microbiol. 70: 4478-4485.
- Ju, W., Cao, G., Rump, L., Strain, E., Luo, Y. and Timme, R., 2012. Phylogenetic analysis of Non-O157 Shiga toxin-producing Escherichia coli strains by whole genome sequencing. J. Clin. Microbiol. 50: 4123-4127.
- Kaper, J. B., Nataro, J. P. and Mobley, H. L., 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2: 123-140.
- Kheiri, R., Ranjbar, R., Khamesipour, F., Akhtari, L., 2016. Role of Antibiotic in Drug Resistance and Integrons Prevalence in Escherichia coli Isolated from Human and Animal Specimens. Kafkas. Univ. Vet. Fak. Derg. 22: 953-959.
- Koczura, R., Mokracka, J., Jablonska, L., Gozdecka, E., Kubek, M. and Kaznowski, A., 2012. Antimicrobial resistance of integron-harboring Escherichia coli isolates from clinical samples, wastewater treatment plant and river water. Sci. Total Environ. 414: 680-685.
- Kümmerer, K., 2009. Antibiotics in the aquatic environment-a review-part I. Chemosphere 75: 417-434.
- Leomil, L., Pestana de Castro, A. F., Krause, G., Schmidt, H. and Beutin, L., 2005. Characterization of two major groups of diarrheagenic Escherichia coli O26 strains which are globally spread in human patients and domestic animals of different species. FEMS Microbiol. Lett. 249: 335-342.
- McLellan, S. L., 2004. Genetic diversity of Escherichia coli isolated from urban rivers and beach water. Appl. Environ. Microbiol. 70: 4658-4665.
- Mendes Silva, D. and Domingues, L., 2015. On the track for an efficient detection of Escherichia coli in water: A review on PCR-based methods. Ecotoxicol. Environ. Saf. 113: 400-411.
- Nataro, J. P. and Kaper, J. B., 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev.11: 142-201.
- National Research Council and Committee on Indicators for Waterborne Pathogens (ed.), 2004. Indicators for waterborne pathogens. National Academies Press, Washington, DC.
- Olive, D. M. and Bean, P., 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37: 1661-1669.
- Olsen, S.J., Miller, G., Breuer, T., Kennedy, M., Higgins, C., Walford, J., McKee, G. and Fox, K., 2002. A waterborne outbreak of Escherichia coli O157:H7 infections and hemolytic uremic syndrome: implications for rural water systems. Emerg. Infect. Dis. 8: 370-375.
- Rademaker, J., Hoste, B., Louws, F., Kersters, K., Swings, J., Vauterin, L., Vauterin, P. and de Bruijn, F., 2000. Comparison of AFLP and rep-PCR genomic fingerprinting with DNA-DNA homology studies: Xanthomonasas a model system. Int. J. Syst. Evol. Microbiol. 50: 665-677.
- Rahimi, E., Khamesipour, F., Yazdi, F. and Momtaz H., 2012. Isolation and characterization of enterohaemorragic Escherichia coli O157: H7 and EHEC O157: NM from raw bovine, camel, water buffalo, caprine and ovine milk in Iran. Kafkas. Univ. Vet. Fak. Derg. 18: 559-564.
- Raissy, M., Khamesipour, F., Rahimi, E. and Khodadoostan, A., 2014. Occurrence of Vibrio spp., Aeromonas hydrophila, Escherichia coli and Campylobacter spp. in crayfish (Astacus leptodactylus) from Iran. IJFS 13: 944-954.
- Ranjbar, R., Hosseini, S., Zahraei-Salehi, T., Kheiri, R. and Khamesipour F., 2016. Investigation on prevalence of Escherichia coli strains carrying virulence genes ipaH, estA, eaeA and bfpA isolated from different water sources. Asian Pac. J. Trop. Dis. 6: 278-283.
- Ranjbar, R., Karami, A., Farshad, S., Giammanco, G.M. and Mammina, C., 2014. Typing methods used in the molecular epidemiology of microbial pathogens: a how-to guide. New Microbiol. 37: 1-15
- Ranjbar, R., Khamesipour, F., Jonaidi-Jafari, N. and Rahimi, E., 2016. Helicobacter pylori in bottled mineral water: genotyping and antimicrobial resistance properties. BMC Microbiol. 16: 1.
- Ranjbar, R., Khamesipour, F., Jonaidi-Jafari, N. and Rahimi, E., 2016. Helicobacter pylori isolated from Iranian drinking water: vacA, cagA, iceA, oipA and babA2 genotype status and antimicrobial resistance properties. FEBS Open Bio. 6: 433-441.
- Sambrook, J., Fritsch, E. F. and Maniatis, T., 1989. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor Laboratory Press, NY. p: 1584.
- Scheutz, F. and Strockbine, N. A., 2005. Genus Escherichia. In Bergey’s Manual of Systematic Bacteriology, 2nd Ed.; Brenner, D. J., Krieg, N. R., Staley, J.T., Eds. Springer, NY, USA, Volume 2, Part B, pp. 607-623.
- Schloter, M., Lebuhn, M., Heulin, T. and Hartmann, A., 2000. Ecology and evolution of bacterial microdiversity. FEMS Microbiol. Rev. 24: 647-660.
- Tajbakhsh, E., Ahmadi, P., Abedpour-Dehkordi, E., Arbab-Soleimani, N. and Khamesipour, F., 2016. Biofilm formation, antimicrobial susceptibility, serogroups and virulence genes of uropathogenic E. coli isolated from clinical samples in Iran. Antimicrob. Resist. Infect. Control 5: 11.
- Tajbakhsh, E., Khamesipour, F., Ranjbar, R. and Ugwu, I. C., 2015. Prevalence of class 1 and 2 integrons in multi-drug resistant Escherichia coli isolated from aquaculture water in Chaharmahal Va Bakhtiari province, Iran. Ann Clin Microbiol Antimicrob 14: 37.
- Torkan, S., Bahadoranian, M. A., Khamesipour, F. and Anyanwu, M. U., 2016. Detection of virulence and antimicrobial resistance genes in Escherichia coli isolates from diarrhoiec dogs in Iran. Arch Med Vet 48: 181-190.
- Vaneechoutte, M., 1996. DNA fingerprinting techniques for microorganisms. A proposal for classification and nomenclature. Mol. Biotechnol. 6: 115-142.
- Versalovic, J., Koeuth, T. and Lupski, J. R., 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic. Acid. Res. 19: 6823-6831.
- Walk, S. T., Alm, E. W., Calhoun, L. M., Mladonicky, J. M. and Whittam, T. S., 2007. Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ. Microbiol. 9: 2274-2288.
- Watkinson, A. J., Micalizzi, G. R., Bates, J. R. and Costanzo, S. D., 2007. Novel method for rapid assessment of antibiotic resistance in Escherichia coli isolates from environmental waters by use of a modified chromogenic agar. Appl. Environ. Microbiol. 7: 2224-2229.
- Yates, M. V., 2007. Classical indicators in the 21st century-far and beyond the coliform. Water Environ. Res. 79, 279-286.