Research Article - Biomedical Research (2018) Volume 29, Issue 1
miR-124-3p and miR-140-3p.2 act as negative regulators of Beclin1 and LC3 expression in the liver of rat model with hepatic impact injury
Yulin Niu1, Yinglian Zhang2, Kun Li2*, Chungen Xing1* and Chengyi Sun3*
1Department of General Surgery, the Second Affiliated Hospital of Soochow University, Suzhou, Jiangsu, 215004, PR China
2Department of Transplantation, the Affiliated Hospital of Guizhou Medical University, 550004, PR China
3Department of Biliary-Hepatic Surgery, the Affiliated Hospital of Guizhou Medical University, Guiyang, Guizhou, 550004, PR China
- *Corresponding Authors:
- Kun Li
Department of Transplantation
The Affiliated Hospital of Guizhou Medical University, PR China
Chungen Xing
Department of General Surgery
The Second Affiliated Hospital of Soochow University, PR China
Chengyi Sun
Department of Biliary-Hepatic Surgery
The Affiliated Hospital of Guizhou Medical University, PR China
Accepted date: June 29, 2017
DOI: 10.4066/biomedicalresearch.29JRG-17-1393
Visit for more related articles at Biomedical ResearchAbstract
Objective: To investigate the effect of autophagy on hepatic impact injury in rats and its molecular mechanism inovlved.
Methods: Sixty healthy male Sprague-Dawley (SD) rats (clean grade) were randomly divided into two groups, the normal group and the model group (n=30 in each group). An impact injury model of rat liver was established by impacting rat liver with 200 kPa impact using BIM-V biological impactor. The enzyme activity of ALT and AST in blood serum was measured by rate method; the changes of TNF-α, IL-8 and IL-2 levels were detected by ELISA; morphological changes were observed by microscope after routine HE staining; the formation of autophagosomes was observed by laser scanning confocal microscopy; protein expression levels were detected by Western blotting and immunohistochemical methods, and the relative expression levels of genes were determined by RT-PCR; Bioinformatics software was used to analysis the regulate miRNAs of microtubule-associated protein l light chain 3 (LC3) and Beclin1; a double-fluorescent reporter gene vector was constructed to detect the interactions between genes.
Results: The serum levels of ALT, AST, TNF, IL-8 and IL-2 in the model group were markedly higher than those in the normal group, and the difference was statistically significant (p<0.01). The morphology of the liver tissue in the normal group was normal, versus that in the model group was damaged to different degrees, for example, the sheet necrosis occurred in the surrounding area of the central veins of hepatic lobules where a large number of necrotic cells were observed. The expression of LC3 and Beclin1 proteins of liver tissues in the model group was significantly increased (p<0.05). MiR-124-3p negatively regulated the Beclin1 expression and miR-140-3p.2 negatively regulated the LC3 expression.
Conclusion: Autophagy plays an important role in the hepatic impact injury in rats, and miR-124-3p and miR-140-3p.2 may act as negative regulators of Beclin1 and LC3 expressions, respectively.
Keywords
Liver impact injury, Autophagy, LC3, Beclin1, MicroRNA
Abbreviations
LC3: Microtubule-Associated Protein 1 Light Chain 3; ROS: Reactive Oxygen Species; ALT: Alanine Aminotransferase; AST: Aspartate Aminotransferase; ATG: Autophagy-Related Gene; RNA: Ribonucleic Acid; HE: Hematoxylin-Eosin; ELISA: Enzyme Linked Immunosorbent Assay; TNF: Tumor Necrosis Factor; IL-8: Interleukin -8; IL-2: Interleukin-2.Introduction
Autophagy, or cellular self-digestion, plays an important role in cell growth and development and is a key way for organelles to maintain the homeostasis [1]. Studies have shown that autophagy could be activated when liver cells were in the state of nutritional deficiencies and liver tissue was under the conditions of ischemia and hypoxia. Inhibition of autophagy can produce a large number of reactive oxygen species (ROS) and induce necrosis in a number of liver cells. When cells lack of nutrition, autophagy provides a protective effect and can promote cell survival. Liver tolerance to ischemic injury in rats increases as autophagy increases [2]. Cell homeostasis would be destroyed when the autophagic clearance of ischemiareperfusione induced damaged mitochondria is impaired. It would change mitochondrial permeability, destroy the coupling effect of oxidative phosphorylation, eventually leading to liver cell death. Many studies have shown that autophagy was closely related to the pathogenesis of various liver diseases, such as viral hepatitis, liver fibrosis, liver cancer and fatty liver disease [3].
MicroRNAs (miRNAs) are a class of conserved, endogenous single-stranded, non-coding small molecule RNAs with about 19 to 25 nucleotides in length (nt). It can cause the degradation of target mRNAs or inhibit protein translation by binding to specific target sites in the 3' untranslated region (3' UTR) of target mRNAs and thus regulate gene expression after transcription, influence individual development, cell apoptosis, cell proliferation and differentiation and other life activities [4]. Cell autophagy is regulated by autophagic genes Beclin1 and LC3 [5]. The analysis using biological software targerscan has shown that Beclin1 was the target gene regulated by miR-124-3p; likewise, LC3 was the target gene regulated by miR-140-3p.2. Therefore, we speculate that the miRNAs also participate in the regulation of autophagy. Currently, there is little research on the autophagy in liver trauma both at demestic and abroad. This study, for the very first time, explored the expression and mechanism of autophagy-related proteins LC3 and Beclin1 in liver tissue after hepatic impact injury.
Methods
Establishment of rat model with hepatic impact injury
The rat model of hepatic impact injury was established for the purposes of a previous unpublished study [6], and the tissue specimens from animals were obtained from the archives of the Affiliated Hospital of Guizhou Medical University with informed consent and with the approval of the institutional ethics committee (Guizhou, China). The methods used to establish this model were as follows: Thirty male Sprague Dawley rats (SD, clean grade).Thirty male Sprague-Dawley (SD, clean grade,) rats, 80 weeks of age, weighing 250 ± 20 g, were purchased from the Animal Experimental Center of Guizhou Medical University (animal batch number, SYXK Qian 2015-0005; ethnic batch number, 1502014). All rats were fed at the Animal Experimental Center of Guizhou Medical University during the experiment. For adaptation, the rats were fed for a week before further manipulation. Animals were housed under standard lighting conditions and were allowed to freely access water. After feeding with normal diet for 1 week at the room temperature, the rats were randomly divided into normal group (group A) and model group (group B). For the model group, the hepathic blunt impact injury was produced by using BIM-V biological impactor with 200KPa impact [7]. After 2 h, the rats were sacrificed by femoral artery bloodletting, 1 ml of blood was extracted and the serum was retained routinely; The rats were not anaesthetized prior to this procedure.at the same time, damage classification were conducted according to literature [8]. Rats in the model group consisted of 3 cases with the lesion in the left liver lobe, 9 cases in the middle liver lobe, 6 cases in both the left and middle liver lobes and 2 cases of simple lacerations. The liver tissue at the same location of all the rats was collected, fixed with 10% neutral formaldehyde and embedded with paraffin and stained with HE. The remaining liver tissue was stored at -80°C. The experiments was performed on June 2016. The ethical approval for the animal experiments performed in the previously unpublished study was obtained from the Ethics Committee of Guizhou University.
Determination of relevant biochemical indicators
Serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) activities were detected in strict accordance with the manufacture’s instructions of the kit using automatic biochemical analyzer. The levels of TNF-α, IL-8 and IL-2 were detected by enzyme linked immunosorbent assay (ELISA), which were performed according to the manufacture’s instruction.
Immunocytochemistry
The liver tissue was fixed at 4°C for 10 min in pre-cooled 100% methanol (-20°C), rinsed with phosphate-buffered saline (PBS) for three times, then permeabilized at room temperature in 0.2%Triton X-100 for 5 min and blocked at room temperature for 30 min in 5% BSA and 0.1% triton X-100. Samples were subsequently incubated with primary antibodies at 4°C overnight, followed by a secondary antibody conjugated to rabbit Alexa Fluor 488 (1:500; Molecular Probes, Eugene, OR, USA) for 1 h. After rinsing for three times with PBS, the cells were incubated with 0.5 ng/ml of DAPI for 15 min at room temperature and then sealed with mounting medium. All samples were examined under a laser scanning confocal microscope (Nikon C1-Si, Mississauga, Canada).
The expression of Beclin1 and LC3 proteins in liver tissue determined by immunohistochemistry assay
Rat liver tissue was fixed with 4% formaldehyde followed by routine dehydration and paraffin embedding. Paraffinembedded tissue was sliced into 4 um sections, followed by dewaxing with xylene and gradient alcohol hydration in strict accordance with the kit instructions (PV-900, Beijing Zhongyu Jinqiao Biotechnology Co., Ltd.). Beclin1 and LC3 antibodies were purchased from Abcam Corporation, UK, and used at 1:1000 dilution. For negative controls, the primary antibodies were replaced with PBS. The results showed that Beclin1 and LC3 were present in both the cytoplasm and nucleus, the positive expression were light yellow to yellowish brown granules , and the areas with brownish yellow color were positive. Five fields were randomly selected for each section (magnification 400X), 100 cells were counted and the percentage of positive cells was calculated to indirectly estimate the expression levels.
Western blot analysis
The liver tissue was lysed in RIPA buffer containing a proteinase inhibitor cocktail (Biocolor BioScience & Technology, Shanghai, China). After centrifugation at 4°C for 15 min, the pellets were discarded. The protein concentration of each group was determined by bicinchoninic acid (BCA) Protein Assay Kit (Bioss, Beijing, China). After preparation of the gel, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed. GAPDH was used as the internal reference. The ratios of Beclinl, LC3 and GAPDH were calculated and interpreted as the relative expressions of Beclinl and LC3 proteins, and all experiments were repeated for three times (absolute gray value=gray valuebackground gray value).
Cell culture
Rat liver derived cell line BRL-3A was purchased from the Shanghai Institute of Cell Biology (Shanghai, China). The cells were cultured in Dulbecco’s modified Eagle medium (Gibco BRL, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS, HyClone, Invitrogen, Camarillo CA, USA), 100 U/ml of penicillin (Invitrogen, Camarillo, CA, USA), and 100μg/ml of streptomycin (Invitrogen, Camarillo, CA, USA). Cells were cultured at 37°C in a humidified incubator in the presence of 5% CO2. All cell lines were passaged for fewer than 6 months.
Luciferase reporter assay
The 3’ UTR of the gene was obtained by PCR amplification and cloned separately into multiple cloning sites of the psi- CHECKTM-2 luciferase miRNA expression reporter vector. BRL-3A cells were transfected with miRNA mimics, miRNA inhibitor, negative control miRNA, a miRNA inhibitor, or empty plasmid (50 nM, Ribobio, Guangzhou, China) using Lipofectamine 2000 according to the manufacturer’s instructions. Nucleotide-substitution mutation analysis was carried out using direct oligomer synthesis of 3’ UTR. All constructs were verified by sequencing. Luciferase activity was measured using the Dual Luciferase Reporter Assay System Kit (Promega, Madison, WI, USA) on a Tecan M200 luminescence reader according to the manufacturer's instructions [9].
Quantitative real-time RT-PCR
Total RNA samples were extracted using Trizol (Invitrogen, Camarillo, CA, USA) according to the manufacturer’s instructions. Real-time quantitative PCR analysis was performed using Applied Biosystems 7500 real-time PCR platforms (Foster City, CA, USA). The U6 was used as an internal control. The primers used in quantitative real-time PCR were shown in Table 1. The levels of gene expression were calculated using the threshold cycle (CT) and 22- DDCtmethod.
| Gene | Primer (5’-3’) |
|---|---|
| miRs (RTprimer) | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTTCCCAT |
| U6 F | 5’-CTCGCTTCGGCAGCACA-3’ |
| U6 R | 5’-AACGCTTCACGAATTTGCGT-3’ |
| hsa-miR-124-3p F | 5’-TCGGCAGGTAAGGCACGCG-3’ |
| hsa-miR-124-3p R | 5’-TCGGCAGGTAAGGCACGCG-3’ |
| hsa-miR-140-3p.2 F | 5’-TCGGCAGGTAACACTGTCTGGT -3’ |
| hsa-miR-140-3p.2 R | 5’-CTCAACTGGTGTCGTGGA-3’ |
Table 1. Primers of genes.
Statistical analysis
All the results were represented the average of at least three independent experiments. Data were expressed as the mean ± SD and analyzed by one-way analysis of variance (ANOVA) and Student's t test. A p<0.05 was considered statistically significant.
Results
Successful establishment of rat model with hepatic blunt impact injury
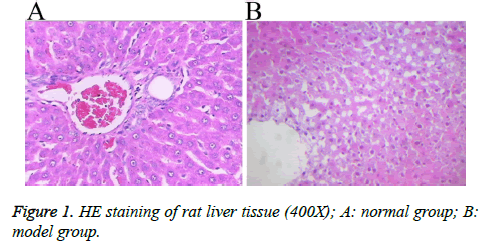
A rat model with hepatic blunt impact injury was established by using BIM-V biological impactor with 200 KPa impact to hit the rat liver. Compared with the normal group, the serum levels of ALT, AST, TNF, IL-8 and IL-2 in the model group were markedly higher than those in the normal group with statistical significance (p<0.01, Table 2). The HE staining result showed that, the morphology of the liver tissue in the normal group was normal, the structure of liver lobules was clear, and liver cells were radially distributed around the central veins. The liver tissue in the model group displayed different degrees of injuries, such as cloudy swelling of liver cells, ballooning degeneration, patchy necrosis surrounding the central veins of the hepatic lobules, focal necrotic area inside the lobule and a large number of necrotic cells (Figure 1). The results showed that the rat model of hepatic blunt impact injury was successfully established.
| Group | n | ALT (U/L) | AST | TNF | IL-8 | IL-2 |
|---|---|---|---|---|---|---|
| a | 15 | 60.29 ± 5.39 | 140.84 ± 4.16 | 38.23 ± 5.42 | 9.6 ± 6.80 | 7.5 ± 6.32 |
| B | 15 | 113.85 ± 9.39a | 194.41 ± 34.86a | 76.38 ± 5.42a | 21.36 ± 6.80a | 31.2 ± 5.70a |
Table 2. Comparison of serum levels of ALT, AST, TNF, IL-8 and IL-2 in two groups (U/L).
Occurrence of autophagy in the liver tissue upon blunt impact injury
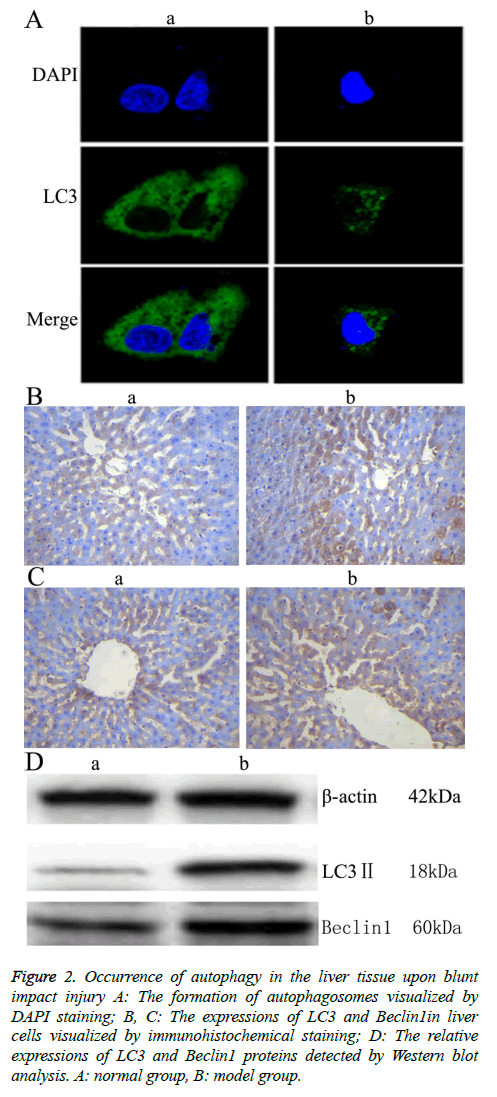
To investigate the autophagic response caused by blunt impact injury in rats, the liver tissue was first stained with DAPI, then incubated with anti-LC3 antibody. Morphologic changes, such as were observed under fluorescence confocal microscopy. The fluorescence is scattered in Figure 2A. The fluorescence is aggregated in Figure 2A. The aggregation indicates autophagy (positive), the scatter indicates no autophagy happened (negative) [10]. Immunohistochemical staining was further conducted, and light yellow to yellowish brown granules in the cytoplasm and nucleus of the liver tissue in the model group indicated the positive expression. The results showed that LC3 and Beclin 1 were mainly expressed in the cytoplasm of hepatocytes, which appeared as brownish yellow area. Immunohistochemical staining of samples from the normal group also showed mild expression of LC3 and Beclin 1. Compared with the normal group, the expressions of LC3 and Beclin1 in liver tissue of the model group were increased significantly (Figures 2B and 2C, Table 3). Moreover, the result of Western blot analysis also indicated that the relative expressions of LC3 and Beclin1 proteins were significantly increased in the model group (Figure 2 D). These results together showed that autophagy might play an important role in rat liver upon blunt impact injury.
| Group | LC3 | Beclin1 |
|---|---|---|
| a | 21.40 ± 2.30 | 18.40 ± 2.96 |
| b | 56.60 ± 5.32* | 49.40 ± 2.70* |
Table 3. Expressions of LC3 and Beclin1 proteins in rat liver tissue (mean ± SD, %).
Figure 2: Occurrence of autophagy in the liver tissue upon blunt impact injury A: The formation of autophagosomes visualized by DAPI staining; B, C: The expressions of LC3 and Beclin1in liver cells visualized by immunohistochemical staining; D: The relative expressions of LC3 and Beclin1 proteins detected by Western blot analysis. A: normal group, B: model group.
MiR-124-3p and miR-141-3p regulated the expressions of target genes Beclin1 and LC3
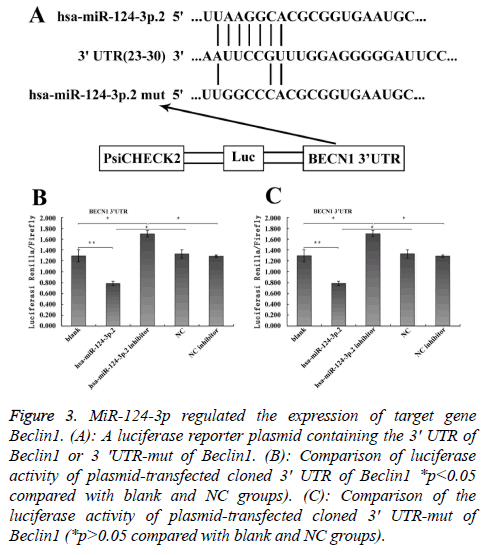
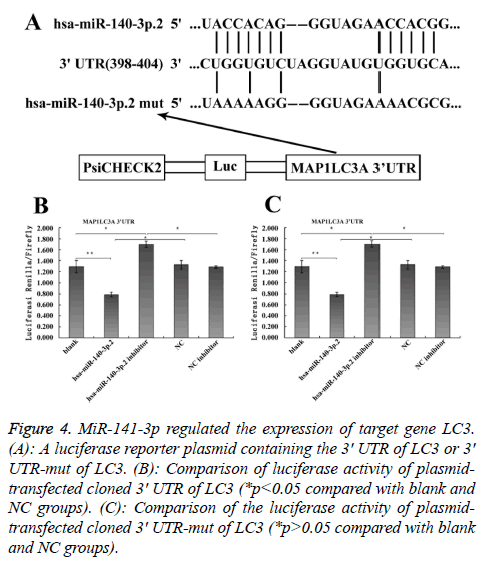
To explore the mechanism involved in the regulation of expression of autophagy-related proteins LC3 and Beclin1 after hepatic blunt injury in rat model, first of all, biological information software (targetscan) was used to predict whether there were binding sites for miR-124-3p and miR-140-3p.2 in the 3' UTR of Beclin1 and LC3, espectively. The predicted miR-124-3p binding sites were at position 23-30 of the Beclin1 3' UTR, and the predicted miR-141-3p binding sites were at position 398-404 of the LC3 3' UTR (Figures 3A and 4A). To determine whether the observed effects depend on the regulation of the 3′ UTR of LC3 and Beclin1, a luciferase reporter plasmid containing the 3′ UTR of LC3 and Beclin1 was constructed. Co-transfection of BRL-3A cells with miR-140-3p.2 and miR-124-3p mimics was performed to confirm the binding site on LC3 and Beclin1 3' UTR. The cotransfection of BRL-3A cells with miR-140-3p.2, miR-124-3p mimics and psiCHECK-2-LC3 3' UTR, psiCHECK-2-Beclin1 3' UTR significantly inhibited luciferase activity (Figures 3B and 4B); however, co-transfection of BRL-3A cells with miR-140-3p.2, miR-124-3p mimics and psiCHECK-2-mut- LC3 3' UTR, psiCHECK-2-mut-Beclin1 3' UTR had little effect on the activity of luciferase (p>0.05, Figures 3C and 4C). The above results demonstrated that miR-140-3p.2 and miR-124-3p played regulatory roles by binding to 3' UTR of LC3 and Beclin 1, respectively.
Figure 3: MiR-124-3p regulated the expression of target gene Beclin1. (A): A luciferase reporter plasmid containing the 3' UTR of Beclin1 or 3 'UTR-mut of Beclin1. (B): Comparison of luciferase activity of plasmid-transfected cloned 3' UTR of Beclin1 *p<0.05 compared with blank and NC groups). (C): Comparison of the luciferase activity of plasmid-transfected cloned 3' UTR-mut of Beclin1 (*p>0.05 compared with blank and NC groups).
Figure 4: MiR-141-3p regulated the expression of target gene LC3. (A): A luciferase reporter plasmid containing the 3' UTR of LC3 or 3' UTR-mut of LC3. (B): Comparison of luciferase activity of plasmidtransfected cloned 3' UTR of LC3 (*p<0.05 compared with blank and NC groups). (C): Comparison of the luciferase activity of plasmidtransfected cloned 3' UTR-mut of LC3 (*p>0.05 compared with blank and NC groups).
MiR-124-3p and miR-141-3p negatively regulated the expressions of target genes Beclin1 and LC3
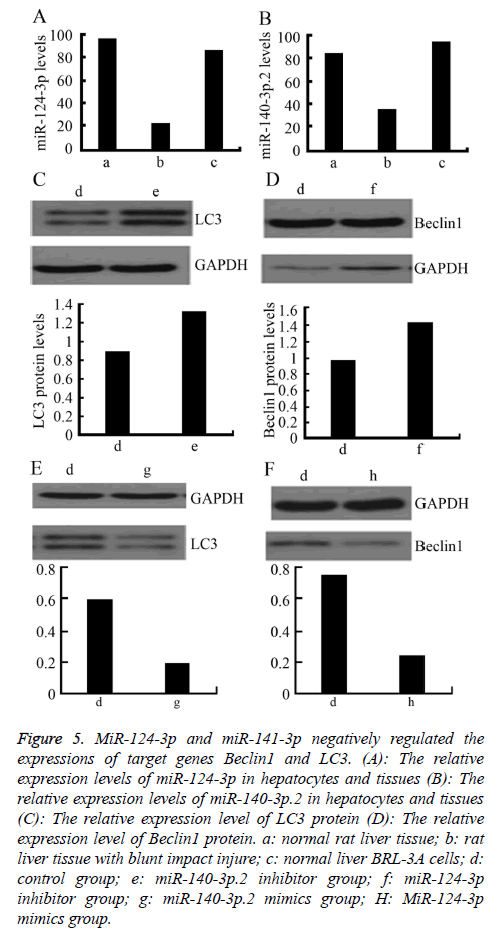
To further explore the regulatory effect of miR-124-3p and miR-141-3p on Beclin1 and LC3, firstly, the relative expression levels of miR-140-3p.2 and miR-124-3p in normal rat liver BRL-3A cells was detected by RT-PCR. The results showed that the expressions of miR-140-3p.2 and miR-124-3p in normal liver tissue were significantly higher than those in the liver tissue with blunt impact injury. Moreover, they were also highly expressed in normal liver BRL-3A cells (Figures 5A and 5B). Western blotting was used to detect the expressions of LC3 and Beclin1 proteins after certain concentrations of miR-140-3p.2 and miR-124-3p inhibitors were transfected into normal liver BRL-3A cells for 48 h. The result showed that the expression levels of LC3 and Beclin1 proteins were significantly upregulated (Figures 5C and 5D). When certain concentrations of miR-140-3p.2 and miR-124-3p mimics were transfected into normal liver BRL-3A cells for 48 hours, the Western blotting results showed that the expression levels of LC3 and Beclin1 proteins were significantly downregulated (Figures 5E and 5F). The above results suggested that miR-124-3p negatively regulated the expression of Beclin1 and miR-140-3p.2 negatively regulated the expression of LC3 in normal liver cells.
Figure 5: MiR-124-3p and miR-141-3p negatively regulated the expressions of target genes Beclin1 and LC3. (A): The relative expression levels of miR-124-3p in hepatocytes and tissues (B): The relative expression levels of miR-140-3p.2 in hepatocytes and tissues (C): The relative expression level of LC3 protein (D): The relative expression level of Beclin1 protein. a: normal rat liver tissue; b: rat liver tissue with blunt impact injure; c: normal liver BRL-3A cells; d: control group; e: miR-140-3p.2 inhibitor group; f: miR-124-3p inhibitor group; g: miR-140-3p.2 mimics group; H: MiR-124-3p mimics group.
Discussion
Autophagy is a highly sensitive process for cells to cope with various stressful environments, allowing cells to maintain substance and energy balance [11]. LC3, a homologue of yeast Atg8, plays an indispensable role in the whole autophagy process, moreover, LC3 II directly reflect the number of selfvesicles. Beclin1 is essential at the initial stage of autophagy. These two proteins are considered as two reliable markers of autophagic activity [12-15]. Both maintenance of liver physiological functions and cell survival require oxygen supply and liver cells are highly sensitive to hypoxia and ischemia. Long-term ischemia and hypoxia easily cause serious acidosis, ultimately leading to liver cell death. Liver ischemia injury often occurs in liver trauma, shock, liver transplantation and so on. Autophagy, as an important participant, is upregulated in the whole process of ischemia [16].
In this study, the BIM-V biological impactor was used to establish the rat model with hepatic blunt impact injury. The formation of autophagosomes was observed under the fluorescence confocal microscopy, and the expressions of LC3 and Beclin1 proteins were shown to be significantly upregulated according to immunohistochemistry and Western blot. These results suggested that autophagic activity was upregulated in the liver tissue of rat model with hepatic impact injury, thus we speculated that autophagic activity was altered in the liver tissue of the rat model of hepatic impact injury. In the model group, the liver of rat was subjected to external force impact. As a result, the body was under energy and nutrition stress state and thus the metabolism was active. Although the blood supply for the liver was abundant, the abilities of decomposing liver glycogen and synthesizing protein were decreased in the stress state, leading to limited energy usage. Our results suggested that autophagy in the liver tissue with hepatic impact injury was increased, which may alleviate the metabolic stress and aid to maintain the cell survival under acute stress.
Autophagy is no longer simply considered as a physiological process, instead it is now known to participate in the regulation of the occurrence and development of various diseases [17]. Therefore, extensive research regarding autophagy mechanism will provide new insights for the future clinical treatment of various diseases. In this study, the expressions of miR-140-3p. 2 and miR-124-3p were downregulated in the liver tissue after hepatic impact injury, and the result from the Luciferase reporter assay further demonstrated that miR-124-3p negatively regulated Beclin1, likewise, miR-140-3p.2 negatively regulated the expression of LC3. It was found that the BECN1 gene, which encoded the Beclin1 protein, was the target gene of miR-30a. The regulatory role of miR-30a for autophagy might occur during the formation of liposomes [18]. MiR-34a might regulate the autophagic activity and cause injury in I/R RTECs by targeting Atg4B [19]. MiR-20a transfection studies showed that it was involved in the regulation of autophagic proteins (LC3-II and ATG16L1) [20]. The downregulation of 146b-5p inhibited the proliferation of TPC1 cells by increasing autophagy [21]. MiRNA-383 could decrease autophagy in patients with acute pancreatitis, wherease the downregulation of miRNA-383 expression could increase autophagy in acute pancreatitis [22]. Based on the results from the previous studies mentioned above, we speculated that microRNAs could regulate the expression of autophagy proteins and thus affect the occurrence and development of diseases.
In conclusion, to the best of the author’s knowledge, the present study is the first to demonstrate that autophagy played an important role in the liver tissue of rat model with hepatic impact injury, and its molecular mechanism might be associated with negative regulation of Beclin1 and LC3 expression by miR-124-3p and miR-140-3p.2, respectively. However, the limitation of this study is that, only the alterations of autophagy-related proteins LC3 and Beclin1 expression after liver injury were examined. Our group will further investigate the regulatory role of autophagy and related proteins at different levels of hepatic impact injuries. Meanwhile, the dynamic observation of autophagic activity changes after liver impact injury will be performed and relevant mechanism will be explored to provide the new insights for the treatment of clinical liver impact injury.
Acknowledgement
The present study was supported by the Science and Technology Cooperation Projects of Guizhou Province (Grant number: Yin Ke He (2015)7309).
References
- Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol 2010; 221: 3-12.
- Wang Y, Singh R, Xiang Y. Macroautophagy and chaperone mediated autophagy are required for hepatocyte resistance to oxidantstress. Hepatology 2010; 52: 266-277.
- Leake I. Alcoholic liver disease:insights into the pathogenesis of alcoholic liver disease. Nat Rev Gastroenterol Hepatol 2013; 58: 314-324.
- Jian-Xiang N, Xing-Kai M, Jian-Jun R. Studied microRNA gene expression in human hepatocellular carcinoma by microRNA microarray techniques. World J Gastroenterol 2015; 21: 12605-126011.
- Xifeng W, Ziyan D, Lingyun L, Minhua S, Yong Y. Beclin 1 and p62 expression in non-small cell lung cancer: relation with malignant behaviors and clinical outcome. Int J Clin Exp Pathol 2015; 8: 10644-10652.
- Jinyi H. Expression of autophagy related protein LC3 and Beclin1 in liver tissue after rat liver injury. Guizhou Med University 2016: 1-37.
- Moore EE, Cogbill TH, Jurkovich GJ. Organ injury scaling: spleen and liver (1994revision). J Traum 1995; 38: 323-324.
- Yunzhi Y, Wang ZG, Liu HP. The development and application of a minitype mulfifunctional bio-impactor. J Biomed Eng 2000; 17: 309-312.
- He JH, Li BX, Han ZP, Zou MX, Wang L, Lv YB, Zhou JB, Cao MR, Li YG, Zhang JZ. Snail-activated long non-coding RNA PCA3 up-regulates PRKD3 expression by miR-1261 sponging, thereby promotes invasion and migration of prostate cancer cells. Tumour Biol 2016; 37: 16163-16165.
- Paulina S. Autophagy: Nuclear autophagy in tumour suppression. Nat Rev Mol Cell Biol 2015; 16: 700-701.
- Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Molecular Cell 2010; 40: 280-293.
- Zhang W, He C. Regulation of plasma membrane receptors by a new autophagy-related BECN/Beclin family member. Autophagy 2014; 10: 1472-1473.
- Rautou PE, Mansouri A, Lebrec D, Durand F, Valla D. Autophagy in liver diseases. J Hepatol 2010; 53: 1123-1125.
- Klionsky DJ, Abdalla FC, Abeliovich H. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8: 445-544.
- Kang R, Zeh HJ, Lotze MT. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 2011; 18: 571-580.
- Bechmann LP, Hannivoort RA, Gerken G, Hotamisligil GS, Trauner M, Canbay A. The interaction of hepatic lipid and glucose metabolism in liver diseases. Hepatol 2012; 56: 952-964.
- Yang L, Xiao L, Chen LX. Research progress of autophagy and pulmonary diseases. Prog Biochem Biophys 2012; 39: 861-868.
- Zhu H, Wu H, Liu X. Regulation of autophagy by a beclin 1 -targeted micro -RNA, miR-30a, in cancer cells. Autophagy 2009; 5: 816-823.
- Liu XJ, Hong Q, Wang Z, Yu YY, Zou Xa, Xu LH. MicroRNA-34a Suppresses autophagy in tubular epithelial cells in acute kidney injury. Am J Nephrol 2015; 42: 168-175.
- Kuo-Ting S, Michael YC, Ming-Gene T, I-Kuan W, Shih-Sheng C, Chi-Yuan L. MicroRNA-20a regulates autophagy related protein-ATG16L1 in hypoxia-induced osteoclast differentiation. Bone 2015; 73: 145-153.
- Shi Y, Lu XB, Liu Y, Fan Y, Yuang Q. Down-regulation of microRNA-146b-5p repress proliferation of thyroid carcinoma cells ay enhancing autophagy activity. Chinese J Exp Surg 2016; 33: 1210-1212.
- Shao-Yue Z, Yong-Ming H, Zheng-Peng Y, Zhuang XM, Dong B. MiRNA-383 regulates the acinar cell autophagy in acute pancreatitis. Progress Modern Biomed 2017; 17: 66-69.