Research Article - Biomedical Research (2018) Volume 29, Issue 1
MicroRNA-18b promotes cell proliferation and invasion in non-small cell lung cancer by directly targeting runt-related transcription factor 3
Lei Zhao#, Yanjun Wang# and Chuanfu Jiang*
Department of Thoracic Surgery, the First Affiliated Hospital of Dalian Medical University, PR China
#These authors contributed equally to this work
- *Corresponding Author:
- Chuanfu Jiang
Department of Thoracic Surgery
The First Affiliated Hospital of Dalian Medical University, PR China
Accepted on August 11, 2017
DOI: 10.4066/biomedicalresearch.29-17-1966
Visit for more related articles at Biomedical ResearchAbstract
Lung cancer is the most common cancer and the leading cause of cancer-related deaths in men and women globally. Increasing evidence has demonstrated that microRNAs (miRNAs) may be critical regulators in cancer-related processes. Therefore, understanding the molecular mechanism between Non-Small Cell Lung Cancer (NSCLC) and abnormal miRNA expression levels may help identify novel diagnostic biomarkers and therapeutic targets for patients with NSCLC. MiRNA-18b (miR-18b) is abnormally expressed in multiple types of human cancers. However, the expression pattern, function and underlying mechanisms of miR-18b in NSCLC have yet to be elucidated. This study aimed to detect the expression levels of miR-18b, explore the regulatory roles of miR-18b in NSCLC and reveal the possible underlying mechanisms of miR-18b involved in this process. Experiments indicated that miR-18b was upregulated in NSCLC tissues and cell lines. MiR-18b downregulation suppressed the proliferation and invasion of NSCLC cells. Runt-related transcription factor 3 (RUNX3) was identified as a direct target of miR-18b in NSCLC by a series of experiments. Moreover, RUNX3 overexpression played the same role as miR-18b under expression in NSCLC cells, by a series of experiments. These findings suggested that miR-18b inhibition may provide a novel effective therapeutic method for patients with NSCLC.
Keywords
Non-small cell lung cancer, microRNA-18b, Runt-related transcription factor 3, Proliferation, Invasion.
Introduction
Lung cancer is the most common cancer and the leading cause of cancer-related deaths in men and women globally [1,2]. This disease may be subdivided into small-cell lung carcinoma and Non-Small-Cell Lung Carcinoma (NSCLC) according to histological characteristics [3]. NSCLC is the most common lung cancer subtype that accounts for approximately 85% of the total number of patients with lung cancer [4]. Patients with localized NSCLC that has not spread to lymph nodes are primarily treated with surgery. Patients with this disease are also subjected to radiotherapy, chemotherapy or a combination of targeted therapies [5,6]. Despite the remarkable progress in treatments, the prognosis of patients with NSCLC remains unsatisfactory with a 5 y overall survival rate of 16% [7] because of late disease presentation, tumour heterogeneities within histological subtypes and relatively limited understanding of NSCLC formation and progression [8,9]. Therefore, the molecular mechanisms underlying the tumourigenesis and tumour development of NSCLC should be further investigated to develop novel and effective therapeutic targets for patients with this fatal disease.
MicroRNAs (miRNAs) are a large family of non-coding, endogenous and short RNA molecules with approximately 20-24 nucleotides in length [10]. miRNAs are known as critical gene regulators that directly interact with the 3’-Untranslated regions (3’-UTRs) of their target genes in a base-pairing mechanism, thereby inhibiting translation and degrading target mRNA [11,12]. It has been reported that over 1000 miRNAs are encoded by the mammalian genome, and all miRNAs likely regulate about one-third of all human genes [13,14]. Numerous miRNAs are also dysregulated in various types of human cancers, such as NSCLC [15], gastric cancer [16], breast cancer [17] and bladder cancer [18]. Abnormally expressed miRNA may serve as tumour suppressors or oncogenes, which regulate a wide range of biological processes, such as proliferation, apoptosis, invasion and metastasis [19-21]. Hence, the investigation of miRNA expression patterns, roles and molecular mechanism may have great potential as a prognostic marker and therapeutic targets for antitumor therapy.
MiR-18b is abnormally expressed in multiple types of human cancers [22-24]. However, the expression pattern, function and underlying mechanisms of miR-18b in NSCLC have yet to be elucidated. This study aimed to detect the expression levels of miR-18b, explore the regulatory roles of miR-18b in NSCLC and reveal the possible underlying mechanisms of miR-18b involved in this process.
Materials and Methods
Tissue samples
This study was approved by the Ethics Committee of The First Affiliated Hospital of Dalian Medical University. Written informed consents were also obtained from the all participants. A total of 46 paired NSCLC tissues and adjacent non-tumor tissues were collected from NSCLC patients who underwent surgery resection at Department of Thoracic Surgery, The First Affiliated Hospital of Dalian Medical University, between July 2015 and April 2017. None of these patients were treated with radiotherapy or chemotherapy prior to surgery. Tissue specimens were immediately frozen in liquid nitrogen and stored at -80°C until further use.
Cell lines and oligonucleotide transfection
Four human NSCLC cell lines (A549, H460, SK-MES-1 and SPC-A1), non-tumorigenic bronchial epithelium BEAS 2B cell line and HEK293T cell line were acquired from the Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). NSCLC and HEK293T cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 1% penicillin/ streptomycin (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in a humid atmosphere containing 5% CO2 at 37°C. BEAS 2B cell line was maintained in LHC 9 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% FBS.
MiR-18b inhibitor and corresponding scramble miRNA inhibitor negative control (NC inhibitor) were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). RUNX3 overexpression plasmid (pcDNA3.1-RUNX3) and empty plasmid (pcDNA3.1) were obtained from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). Cell transfection was performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissues or cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The concentration of total RNA was determined using the ND-1000 NanoDrop spectrophotometer (NanoDrop Technologies; Shanghai Sangon Co., Ltd.). To quantify miR-18b expression, the total RNA was reverse transcribed into cDNA by using a TaqMan® MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Quantitative polymerase chain reaction (qPCR) was performed with TaqMan MicroRNA Assay kit (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). For RUNX3 mRNA expression, cDNA was synthesized using a Moloney Murine Leukemia Virus Reverse Transcription system (Promega Corporation, Madison, WI, USA), followed by qPCR with a SYBR premix Ex TaqTM kit (Takara Biotechnology Co., Ltd., Dalian, China). U6 and GAPDH were used as an endogenous control for miR-18b and RUNX3 mRNA, respectively. Each sample was analysed in triplicate. The relative expression of miRNA and mRNA was analysed with the 2-ΔΔCt method [25].
Cell Counting Kit 8 (CCK8) assay
Cell proliferation was determined using the CCK8 assay according to the manufacturer’s protocol. Transfected cells were collected at 24 h post-transfection, and seeded into 96- well plates at a density of 3000 cells/well. After 0, 24, 48, or 72 h incubation at 37°C with 5% CO2, 10 μl of CCK8 reagent (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was added into each well. Cells were incubated at 37°C with 5% CO2 for an additional 2 h. The value of absorbance was measured at a wavelength of 450 nm using an ELISA reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Transwell invasion assay
Transwell chambers coated with Matrigel (Becton-Dickinson, Franklin Lakes, NJ, USA) were used to evaluate cell invasion ability. After 48 h of transfection, cells were harvested, suspended in FBS-free DMEM and seeded into the upper compartment of the chamber at a density of 5 × 104 cells. A total of 500 μl DMEM containing 20% FBS was placed in the chamber. Following incubation at 37°C with 5% CO2 for 24 h, the non-invasive cells were removed with cotton swabs. The invasive cells were fixed with 100% methanol, stained with 0.5% crystal violet, washed with PBS and dried in air. The number of invasive cells were photographed and counted using an inverted light microscope (CKX41; Olympus Corporation, Tokyo, Japan; magnification, X200).
Target prediction of miRNAs
To predict miRNAs/mRNAs interactions, TargetScan (www.targetscan.org) and miRBase (http://www.mirbase.org/) were used.
Luciferase report assay
Luciferase reporter plasmid, pGL3-RUNX3-3’-UTR-Wild type (Wt) and pGL3-RUNX3-3’-UTR-Mutant (Mut), were synthesized by Shanghai GenePharma Co., Ltd. HEK293T cells were seeded into 24-well plates at a density of 50-60% confluence. After incubation overnight, HEK293T cells were transfected with luciferase reporter plasmid, along with miR-18b inhibitor or NC inhibitor using Lipofectamine 2000. At 48 h post-co-transfection, firefly and renilla luciferase activities were measured using the Dual-Luciferase assay system (Promega, Madison, WI, USA). Renilla luciferase activity was normalized to firefly luciferase activity. All experiments were performed with three independent replicates.
Western blotting analysis
Total protein was isolated from tissues or cells using radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Haimen, China). Total protein concentration was measured using the Bicinchoninic Acid Protein assay kit (Thermo Fisher Scientific, Inc., Rockford, IL, USA). Equal amounts of protein were separated in 10% SDS polyacrylamide gel electrophoresis, transferred to Polyvinylidene Fluoride (PVDF) membranes (Millipore, Bedford, MA, USA) and blocked with 5% nonfat dried milk in TBS containing 0.05% Tween-20 (TBST) at room temperature for 2 h. Afterwards, the membranes were incubated overnight with the following primary antibodies: mouse anti-human monoclonal RUNX3 antibody (sc-376591; 1:1000 dilution; Santa Cruz Biotechnology, CA, USA) and GAPDH (1:10,00 dilution; sc-47724, Santa Cruz Biotechnology, CA, USA), at 4°C overnight. Subsequently, the membranes were washed three times with TBST and incubated with their respective horseradish peroxidase-conjugated secondary antibody (1:5,000 dilution; sc-2005; Santa Cruz Biotechnology, CA, USA) at room temperature for 2 h. Following three washes in TBST, the protein bands were visualized using an enhanced chemiluminescence solution (Pierce, Rockford, IL, USA). Band intensities were analysed with ImageJ 1.49 (National Institutes of Health, Bethesda, MD).
Statistical analysis
The results were expressed as the mean ± Standard Deviation (SD). SPSS 17.0 (SPSS, Inc., Chicago, IL, USA) was used for statistical analysis with Student's t-tests or one-way ANOVA. SNK (Student-Newman-Keuls) was used to compare between two groups in multiple groups study. In all data analyses, P<0.05 was considered statistically significant.
Results
MiR-18b expression is upregulated in NSCLC tissues and cell lines
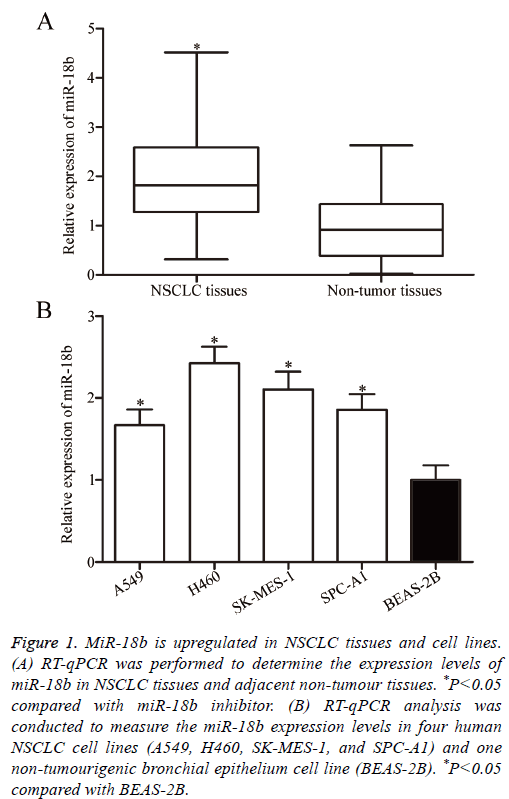
To clarify the biological roles of miR-18b in NSCLC progression, RT-qPCR was performed to measure the miR-18b expression level in 46 paired NSCLC tissues and adjacent nontumour tissues. In Figure 1A, miR-18b was significantly upregulated in NSCLC tissues compared with that in adjacent non-tumour tissues (P<0.05). The miR-18b expression levels were determined in four human NSCLC cell lines, namely, A549, H460, SK-MES-1 and SPC-A1, and one nontumourigenic bronchial epithelium cell line, namely, BEAS-2B. RT-qPCR confirmed that the miR-18b expression was higher in the four NSCLC cell lines than in BEAS-2B (Figure 1B, P<0.05). These results suggested that miR-18b may be involved in NSCLC initiation and progression.
Figure 1: MiR-18b is upregulated in NSCLC tissues and cell lines. (A) RT-qPCR was performed to determine the expression levels of miR-18b in NSCLC tissues and adjacent non-tumour tissues. *P<0.05 compared with miR-18b inhibitor. (B) RT-qPCR analysis was conducted to measure the miR-18b expression levels in four human NSCLC cell lines (A549, H460, SK-MES-1, and SPC-A1) and one non-tumourigenic bronchial epithelium cell line (BEAS-2B). *P<0.05 compared with BEAS-2B.
MiR-18b under expression inhibits the proliferation and invasion of NSCLC cells
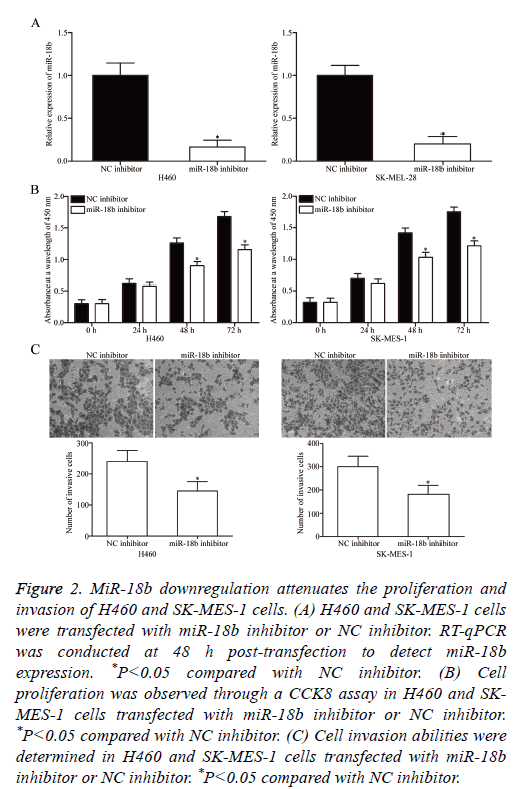
To elucidate the potential effects of miR-18b on the tumourigenicity of NSCLC cells, miR-18b inhibitor or NC inhibitor was transfected into H460 and SK-MES-1 cells, which expressed a relatively higher miR-18b expression level. After transfection was completed, RT-qPCR was conducted to evaluate the transfection efficiency. RT-qPCR data indicated that miR-18b was downregulated in H460 and SK-MES-1 cells transfected with miR-18b inhibitor (Figure 2A, P<0.05).
Figure 2: MiR-18b downregulation attenuates the proliferation and invasion of H460 and SK-MES-1 cells. (A) H460 and SK-MES-1 cells were transfected with miR-18b inhibitor or NC inhibitor. RT-qPCR was conducted at 48 h post-transfection to detect miR-18b expression. *P<0.05 compared with NC inhibitor. (B) Cell proliferation was observed through a CCK8 assay in H460 and SKMES- 1 cells transfected with miR-18b inhibitor or NC inhibitor. *P<0.05 compared with NC inhibitor. (C) Cell invasion abilities were determined in H460 and SK-MES-1 cells transfected with miR-18b inhibitor or NC inhibitor. *P<0.05 compared with NC inhibitor.
The effect of miR-18b under expression on NSCLC cell proliferation was assessed by CCK8 assay. In Figure 2B, miR-18b downregulation inhibited the proliferation of H460 and SK-MES-1 cells (P<0.05). The effects of miR-18b on the invasion abilities of NSCLC cells were investigated through Transwell invasion assay, and the results revealed that miR-18b knockdown markedly decreased the invasion capacities of H460 and SK-MES-1 cells (Figure 2C, P<0.05). These results suggested that miR-18b may play a crucial role in NSCLC cell growth and metastasis.
RUNX3 is a direct target of miR-18b in NSCLC
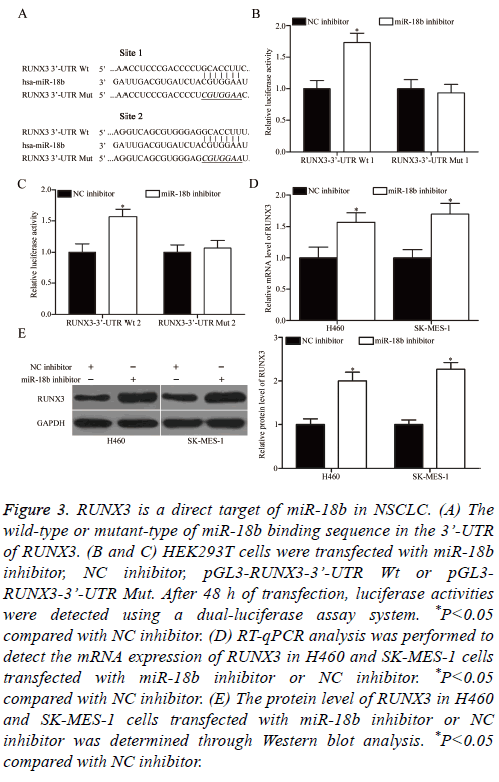
To understand the mechanisms responsible for the oncogenic roles of miR-18b in NSCLC, bioinformatics analysis was performed to search for the possible targets of miR-18b. Among these targets, RUNX3 was selected for further confirmation because it exhibits weak expression in NSCLC and contributes to NSCLC occurrence and development [26-29]. Two high-scoring binding sites of miR-18b were found on the 3’-UTR of RUNX3 (Figure 3A). To confirm this hypothesis, we conducted luciferase reporter assays and explored whether miR-18b can directly target the 3’-UTR of RUNX3. MiR-18b inhibitor or NC inhibitor was transfected into HEK293T cells, together with pGL3-RUNX3-3’-UTR Wt (1 and 2) or pGL3-RUNX3-3’-UTR Mut (1 and 2). In Figures 3B and 3C, the transfection of miR-18b inhibitor significantly upregulated the luciferase activities when this inhibitor was cotransfected with the wild-type 3’-UTR of RUNX3 (1 and 2) in H460 and SK-MES-1 cells (P<0.05). Conversely, the regulatory effect of miR-18b on luciferase activities was abolished after the inhibitor was co-transfected with the mutant 3’-UTR of RUNX3 (1 and 2). We also explored whether miR-18b can regulate the endogenous RUNX3 expression in NSCLC. RT-qPCR and Western blot analysis were conducted to detect the mRNA and protein expression of RUNX3 in H460 and SK-MES-1 cells transfected with miR-18b inhibitor or NC inhibitor. The results showed that the miR-18b downregulation increased the mRNA (Figure 3D, P<0.05) and protein (Figure 3E, P<0.05) expression levels of RUNX3 in H460 and SK-MES-1 cells. These results suggested that RUNX3 is a direct target of miR-18b in NSCLC.
Figure 3: RUNX3 is a direct target of miR-18b in NSCLC. (A) The wild-type or mutant-type of miR-18b binding sequence in the 3’-UTR of RUNX3. (B and C) HEK293T cells were transfected with miR-18b inhibitor, NC inhibitor, pGL3-RUNX3-3’-UTR Wt or pGL3- RUNX3-3’-UTR Mut. After 48 h of transfection, luciferase activities were detected using a dual-luciferase assay system. *P<0.05 compared with NC inhibitor. (D) RT-qPCR analysis was performed to detect the mRNA expression of RUNX3 in H460 and SK-MES-1 cells transfected with miR-18b inhibitor or NC inhibitor. *P<0.05 compared with NC inhibitor. (E) The protein level of RUNX3 in H460 and SK-MES-1 cells transfected with miR-18b inhibitor or NC inhibitor was determined through Western blot analysis. *P<0.05 compared with NC inhibitor.
Upregulation of RUNX3 imitates the roles of miR-18b in NSCLC
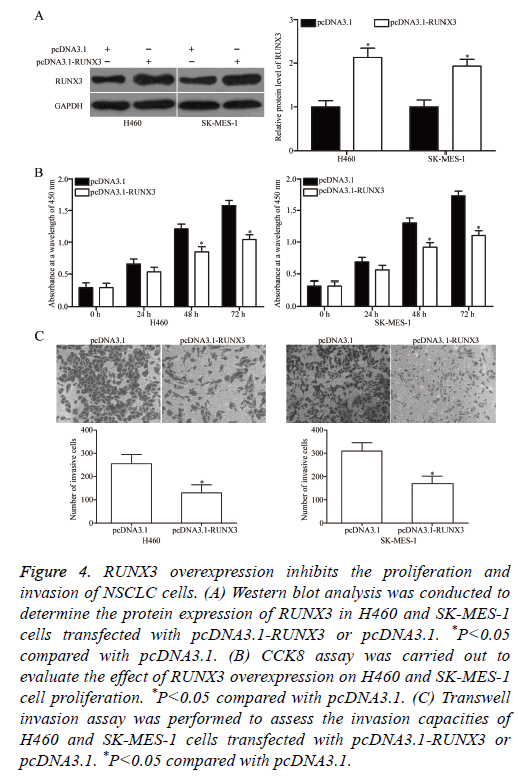
RUNX3 was validated as a direct target of miR-18b in NSCLC. Hence, we hypothesized that the biological roles of RUNX3 overexpression on NSCLC cells are similar to those induced by miR-18b under expression. To confirm this hypothesis, we transfected H460 and SK-MES-1 cells with pcDNA3.1-RUNX3 or pcDNA3.1. Western blot analysis revealed that RUNX3 was significantly upregulated in H460 and SK-MES-1 cells transfected with pcDNA3.1-RUNX3 (Figure 4A, P<0.05). Functional experiments demonstrated that RUNX3 overexpression attenuated the proliferation (Figure 4B, P<0.05) and invasion (Figure 4C, P<0.05) of H460 and SK-MES-1 cells. These results supported the hypothesis that RUNX3 overexpression plays the same role as miR-18b under expression in NSCLC cells and further suggested that RUNX3 may be a functional target of miR-18b in NSCLC.
Figure 4: RUNX3 overexpression inhibits the proliferation and invasion of NSCLC cells. (A) Western blot analysis was conducted to determine the protein expression of RUNX3 in H460 and SK-MES-1 cells transfected with pcDNA3.1-RUNX3 or pcDNA3.1. *P<0.05 compared with pcDNA3.1. (B) CCK8 assay was carried out to evaluate the effect of RUNX3 overexpression on H460 and SK-MES-1 cell proliferation. *P<0.05 compared with pcDNA3.1. (C) Transwell invasion assay was performed to assess the invasion capacities of H460 and SK-MES-1 cells transfected with pcDNA3.1-RUNX3 or pcDNA3.1. *P<0.05 compared with pcDNA3.1.
Discussion
Increasing evidence has demonstrated that miRNAs may be critical regulators in cancer-related processes [30,31]. Therefore, understanding the molecular mechanism between NSCLC and abnormal miRNA expression levels may help identify novel diagnostic biomarkers and therapeutic targets for patients with NSCLC. In this study, miR-18b was upregulated in NSCLC tissues and cell lines, and the downregulated miR-18b inhibited the proliferation and invasion of NSCLC cells. RUNX3 was also identified as a novel direct target gene of miR-18b in NSCLC, and RUNX3 overexpression played the same role as miR-18b under expression in NSCLC cells. These findings suggested that miR-18b may be a novel target for the therapy of patients with NSCLC.
Previous studies have demonstrated that miR-18b is abnormally expressed in multiple types of human cancer. For example, miR-18b is weakly expressed in melanoma tissues, and its low expression is significantly associated with the thickness and stage of melanoma [23]. However, miR-18b expression is upregulated in breast cancer tissues and cell lines [24]. miR-18b expression is also significantly higher in poorly differentiated hepatocellular carcinoma tissues than in welldifferentiated hepatocellular carcinoma. The relapse-free period of patients with hepatocellular carcinoma and high miR-18b expression is significantly shorter than that of patients with the same disease and low miR-18b expression [22]. These findings suggested that the expression pattern of miR-18b exhibits tissue specificity and this miRNA may be developed as a diagnostic biomarker for these types of human cancers.
MiR-18b is identified as a tumour suppressor in several types of human cancers. For instance, Chen et al. reported that miR-18b upregulation inhibits melanoma cell proliferation, colony formation and glycolysis, induces cell cycle arrest in the G1 stage in vitro and reduces tumour growth in vivo [23,32,33]. Nevertheless, miR-18b is identified as an oncogene that accelerates breast cancer cell migration [24]. Murakami et al. found that ectopic miR-18b expression promotes cell proliferation and decreases cell adhesion ability in hepatocellular carcinoma [22]. These conflicting findings indicated that the functions of miR-18b in tumours are diverse and tissue specific because of the non-ideal complementarity of the interactions between miRNAs and their direct target genes. These findings also suggested that miR-18b is a promising molecular target for the treatment of these diseases.
Several direct miR-18b targets, including HIF-1α [23], MDM2 [32,33] in melanoma and TNRC6B [22] in hepatocellular carcinoma, have been validated. In this study, RUNX3 was demonstrated as a direct target of miR-18b in NSCLC. RUNX3 is downregulated in multiple types of human cancers, including ovarian cancer [34], bladder cancer [35], prostate cancer [36], breast cancer [37], gastric cancer [38] and colorectal cancer [39]. Functional studies have demonstrated that RUNX3 serves as a tumour suppressor that inhibits cell proliferation, colony formation, migration, invasion and angiogenesis and promotes cell cycle arrest and apoptosis [40-43]. In NSCLC, RUNX3 expression levels are lower in tumour tissues [26] and may be associated with promoter hypermethylation [44]. Low RUNX3 levels are correlated with poorly differentiated types in patients with NSCLC [26]. The 5-year survival rate of patients with NSCLC and high RUNX3 levels is higher than that of patients with low miR-18b expression levels [26]. RUNX3 participates in NSCLC formation and progression through the regulation of cell proliferation [27], epithelial-mesenchymal transition [28] and tumourigenesis [29]. Combining previous findings with our results, we recommend that the miR-18b/RUNX3 pathway should be further investigated as a therapeutic strategy for patients with NSCLC.
In conclusion, this study demonstrated that miR-18b was significantly upregulated in NSCLC. In vitro studies revealed that miR-18b under expression suppressed the proliferation and invasion of NSCLC cells. RUNX3 was mechanistically identified as a direct target gene of miR-18b in NSCLC, and miR-18b might be a novel target for the therapy of patients with NSCLC.
References
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin 2010; 60: 277-300.
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87-108.
- Travis WD. Pathology of lung cancer. Clin Chest Med 2011; 32: 669-692.
- Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc 2008; 83: 584-594.
- Lazzari C, Bulotta A, Ducceschi M. Historical evolution of second-line therapy in non-small cell lung cancer. Front Med (Lausanne) 2017; 4: 4.
- Jiang T, Qiao M, Zhou F, Ren S, Su C, Zhou C. Effect of combined therapy inhibiting EGFR and VEGFR pathways in non-small-cell lung cancer on progression-free and overall survival. Clin Lung Cancer 2017; 18: 421-431.
- Moumtzi D, Lampaki S, Zarogoulidis P. Prognostic factors for long term survival in patients with advanced non-small cell lung cancer. Ann Transl Med 2016; 4: 161.
- Simon J. Technology radiation technology targets tumors. Surgical precision without the incision. SD Med 2014; 67: 362.
- Johtatsu T, Noguchi S, Yatera K. A case of lung adenocarcinoma with uncontrollable myocardial metastasis and pericardial effusion. J UOEH 2014; 36: 199-203.
- Su Z, Yang Z, Xu Y, Chen Y, Yu Q. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget 2015; 6: 8474-8490.
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136: 215-233.
- Monroig PC, Chen L, Zhang S, Calin GA. Small molecule compounds targeting miRNAs for cancer therapy. Adv Drug Deliv Rev 2015; 81: 104-116.
- Berezikov E, van Tetering G, Verheul M. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res 2006; 16: 1289-1298.
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120: 15-20.
- He M, Xue Y. MicroRNA-148a suppresses proliferation and invasion potential of non-small cell lung carcinomas via regulation of STAT3. Onco Targets Ther 2017; 10: 1353-1361.
- Pan T, Chen W, Yuan X, Shen J, Qin C, Wang L. miR-944 inhibits metastasis of gastric cancer by preventing the epithelial-mesenchymal transition via MACC1/Met/AKT signaling. FEBS Open Bio 2017; 7: 905-914.
- Yao J, Zhang P, Li J, Xu W. MicroRNA-215 acts as a tumor suppressor in breast cancer by targeting AKT serine/threonine kinase 1. Oncol Lett 2017; 14: 1097-1104.
- Wei Z, Hu X, Liu J, Zhu W, Zhan X, Sun S. MicroRNA-497 upregulation inhibits cell invasion and metastasis in T24 and BIU-87 bladder cancer cells. Mol Med Rep 2017; 16: 2055-2060.
- Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med 2014; 20: 460-469.
- Lu YJ, Liu RY, Hu K, Wang Y. MiR-541-3p reverses cancer progression by directly targeting TGIF2 in non-small cell lung cancer. Tumour Biol 2016; 37: 12685-12695.
- Yang S, Zhang Y, Zhao X, Wang J, Shang J. microRNA-361 targets Wilms tumor 1 to inhibit the growth, migration and invasion of non-small-cell lung cancer cells. Mol Med Rep 2016; 14: 5415-5421.
- Murakami Y, Tamori A, Itami S. The expression level of miR-18b in hepatocellular carcinoma is associated with the grade of malignancy and prognosis. BMC Cancer 2013; 13: 99.
- Chen Y, Zhang Z, Luo C, Chen Z, Zhou J. MicroRNA-18b inhibits the growth of malignant melanoma via inhibition of HIF-1alpha-mediated glycolysis. Oncol Rep 2016; 36: 471-479.
- Fonseca-Sanchez MA, Perez-Plasencia C, Fernandez-Retana J. microRNA-18b is upregulated in breast cancer and modulates genes involved in cell migration. Oncol Rep 2013; 30: 2399-2410.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402-408.
- Araki K, Osaki M, Nagahama Y. Expression of RUNX3 protein in human lung adenocarcinoma: implications for tumor progression and prognosis. Cancer Sci 2005; 96: 227-231.
- Torshabi M, Faramarzi MA, Tabatabaei Yazdi M, Ostad SN, Gharemani MH. Runx3 expression inhibits proliferation and distinctly alters mRNA expression of Bax in AGS and A549 cancer cells. Iran J Pharm Res 2011; 10: 355-361.
- Lee JM, Shin JO, Cho KW. Runx3 is a crucial regulator of alveolar differentiation and lung tumorigenesis in mice. Differentiation 2011; 81: 261-268.
- Lee KS, Lee YS, Lee JM. Runx3 is required for the differentiation of lung epithelial cells and suppression of lung cancer. Oncogene 2010; 29: 3349-3361.
- Wang J, Shi C, Wang J, Cao L, Zhong L, Wang D. MicroRNA-320a is downregulated in non-small cell lung cancer and suppresses tumor cell growth and invasion by directly targeting insulin-like growth factor 1 receptor. Oncol Lett 2017; 13: 3247-3252.
- Volinia S, Calin GA, Liu CG. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 2006; 103: 2257-2261.
- Dar AA, Majid S, Rittsteuer C. The role of miR-18b in MDM2-p53 pathway signaling and melanoma progression. J Natl Cancer Inst 2013; 105: 433-442.
- Jazirehi AR, Torres-Collado AX, Nazarian R. Role of miR-18b/MDM2/p53 circuitry in melanoma progression. Epigenomics 2013; 5: 254.
- Lee CW, Chuang LS, Kimura S. RUNX3 functions as an oncogene in ovarian cancer. Gynecol Oncol 2011; 122: 410-417.
- Dodurga Y, Avci CB, Satiroglu-Tufan NL. Detection of deleted in malignant brain tumors 1 and runt-related transcription factor 3 gene expressions in bladder carcinoma. Mol Biol Rep 2012; 39: 4691-4695.
- Chen F, Wang M, Bai J. Role of RUNX3 in suppressing metastasis and angiogenesis of human prostate cancer. PLoS One 2014; 9: 86917.
- Jiang Y, Tong D, Lou G, Zhang Y, Geng J. Expression of RUNX3 gene, methylation status and clinicopathological significance in breast cancer and breast cancer cell lines. Pathobiology 2008; 75: 244-251.
- Hsu PI, Hsieh HL, Lee J. Loss of RUNX3 expression correlates with differentiation, nodal metastasis, and poor prognosis of gastric cancer. Ann Surg Oncol 2009; 16: 1686-1694.
- Zheng W, Zheng K, Zhong L, Li Q, Huang Z. Expression of Runx3 and C-myc in human colorectal cancer. Nan Fang Yi Ke Da Xue Xue Bao 2014; 34: 1042-1047.
- Jili S, Eryong L, Lijuan L, Chao Z. RUNX3 inhibits laryngeal squamous cell carcinoma malignancy under the regulation of miR-148a-3p/DNMT1 axis. Cell Biochem Funct 2016; 34: 597-605.
- Chen F, Liu X, Cheng Q, Zhu S, Bai J, Zheng J. RUNX3 regulates renal cell carcinoma metastasis via targeting miR-6780a-5p/E-cadherin/EMT signaling axis. Oncotarget 2016; 1-16.
- Kim BR, Kang MH, Kim JL. RUNX3 inhibits the metastasis and angiogenesis of colorectal cancer. Oncol Rep 2016; 36: 2601-2608.
- Li H, Li D, Meng N. Effects of RUNX3 mediated Notch signaling pathway on biological characteristics of colorectal cancer cells. Int J Oncol 2017; 50: 2059-2068.
- Lee YS, Lee JW, Jang JW. Runx3 inactivation is a crucial early event in the development of lung adenocarcinoma. Cancer Cell 2013; 24: 603-616.