Research Article - Environmental Risk Assessment and Remediation (2018) Volume 2, Issue 3
Microbiological Analysis for Hydrocarbon Exploration
Syed Mohsin Waheed*, Palak Sangal, Shivangi ShuklaDepartment of Biotechnology, Graphic Era University, Dehradun, Uttarakhand, India, 248001
- *Corresponding Author:
- Syed Mohsin Waheed
Department of Biotechnology, Graphic Era University
Dehradun, Uttarakhand, 248001, India
Tel: 9811968019
E-mail: syedmohsinwaheed@yahoo.com
Accepted date: June 12, 2018
Citation: Waheed SM, Sangal P, Shukla S. Microbiological analysis for hydrocarbon exploration. Environ Risk Assess Remediat. 2018;2(3):1-7
Abstract
Search for crude oil and gas is a national priority and the economy of a country greatly depends on it. Various types of surveys are being carried out to find the accumulations of hydrocarbons. The most commonly used conventional surveys are geological and seismic. Nevertheless all oil companies resort to unconventional methodologies termed as surface prospecting techniques. Surface prospecting reduces the risk and enhances success ratio. Geomicrobial prospecting for hydrocarbon is one such methodology which is found highly target focussing. The Geomicrobial surface prospecting technique is a frontier technology used for prospecting hydrocarbon fields. Attempts have been made to exploit the ability of microbes to utilize gaseous hydrocarbons in prospecting for Hydrocarbon deposits. Gaseous hydrocarbons are usually associated with oil deposits and microbes can use these hydrocarbons. It is assumed that soil samples containing gaseous hydrocarbon-utilizing microorganism may be indicative of nearby hydrocarbon accumulations. This indirect method could hopefully increase the chances of finding hydrocarbon reservoir and could be a relatively inexpensive and environment friendly way of discovering areas for hydrocarbon prospective techniques, provides additional exploratory leads for hydrocarbon exploration.
Keywords
Hydrocarbons, Microorganism, Environment, Metabolism
Introduction
Methane is found in seepages associated with oil deposits. The bacteria that can use methane as sole carbon and energy source are rather specific in this ability. However, methane is also generated in nature as a metabolic product in much anaerobic decomposition of organic substrates. The possibility of the anaerobic production of methane would have to be eliminated before the presence of methane specific microbes could consider a reliable indicator of an oil deposit.
A more specific method that eliminates the possibility of hydrocarbons being biological origin is to analyse for the presence of propane and butane oxidizing bacteria. Since these gases are not produced in nature to any great extent form microbiological processes, so it would seem that the presence of these bacteria is a better indicator of oil deposits.
The present studies have been conducted to detect and enumerate the indicator microbes with special reference to propane and butane oxidizing bacteria for hydrocarbon exploration from sub surface sample collected by ONGC scientists during a Geomicrobial survey for hydrocarbon exploration.
The objective of this project is to detect and enumerate the indicator microbes with special reference to propane oxidizing bacteria and butane oxidizing bacteria for hydrocarbon exploration from surface soil samples.
Literature Review
Gaseous hydrocarbon are widespread in nature and numerous types of microbes have evolved that can oxidize these reduced compounds for their sole source of carbon and energy. The use of gaseous carbon source for cell growth is common and found in autotrophic organism. CO2 is reduced and assimilated by the cells as the primary carbon source. However, using reduced forms of carbon such as hydrocarbon, which require oxidation for energy generative metabolism, is less common.
Microbes live essentially in an aqueous milieu; the gaseous hydrocarbon to be substrates must be soluble in water for uptake and metabolism of cell. Thus solubility of the gas in water becomes important determining the availability of the hydrocarbon as a substrate for microbial metabolism (Table 1).
| Hydrocarbon | Solubility (g/106g of water) |
|---|---|
| Methane | 24.4 + 1.0 |
| Ethane | 60.4 + 1.3 |
| Propane | 62.4 + 2.1 |
| n-Butane | 61.4 + 2.6 |
Table 1. Solubility of various gaseous hydrocarbons in water.
Once the gaseous hydrocarbon is dissolved in the aqueous phase and comes in contact with a microorganism, it can then be assimilated. Since gaseous hydrocarbon is lipid soluble, its uptake and transport through cell membrane can be readily accomplished.
About bacteria
Bacteria have been around on this planet for something in the region of 3.5 billion years in some shape or form. The age of the Earth is about 4.5 billion years old and for 3 billion years single celled organisms have ruled the roost. Through their unique adaptability to different biomes, they have over the ages transformed the Earth along with plate tectonics and other natural phenomena, into a place where large multi celled organism can endure for millions of years. The suns energy output must have been fairly constant over this period in order for life to have survived for so long. Because of their size and almost transparency, bacteria are very difficult to observe with an ordinary light microscope and therefore need to be either stained with a dye, or some form of optical enhancement needs to be employed. Staining bacteria with the use of dyes has been around for over a 150 years and has proved to be an excellent tool for identification, especially for medical diagnostic use. The gram stain, which was developed by a bacteriologist called Hans Gram over a century ago is still the main stain used in microbiology. It works by staining the cell cytoplasm either purple (gram positive) or pink (gram negative).
There are two types of single cells on earth today; prokaryotes, which include bacteria (cyanophytes) and Achaea and the eukaryotes, which include the fungi, animals, algae, protozoa and the larger plants. The bacteria are prokaryotes which do not have a membrane surrounded nucleus; their DNA is not surrounded by a membrane but left loose and irregular and does not have Histones (proteins) associated with the DNA. There are no membrane bound organelles and their cell wall is made up from peptidoglycan which is a complex polysaccharide. Bacteria divide by splitting into two parts, each daughter cell identical to the other, this is called binary fission. Bacteria are also of a much smaller size than eukaryotic cells. Prokaryotic cells ruled the earth for something like 3 billion years until they gave rise to the much larger and more complex cells of the eukaryote.
The bacteria that humans are mostly concerned with for obvious reasons are those that cause disease and also the ones that attack our crops. Bacteria are part of us, in fact there are more bacterial cells within and on the human body than there are human cells. Many of the bacteria found living within us are of paramount importance to our wellbeing, producing vitamins and keeping other more pathogenic bacteria at bay. It is only when microbes enter those parts of the human body that they do not belong that problems with disease begin to appear.
The human mouth is home to something in the region of 300 species of bacteria, with a few species of fungi and protozoa thrown in for good luck. The mouth will be teeming with microbes, possibly hundreds of millions. Some species of bacteria are called flagella, means of moving from one part of the environment to another. There are four different arrangements of flagella which can go from a single flagellum to the cell being completely covered with flagella. The flagella are extremely complex and consist of three major regions: basal body, hook and filament. It is only in the later part of the last century that scientists began to get some ideas as to how this mechanism worked. Flagella are very difficult to observe under the light microscope and it is usual for microbiologist to use special stains which adhere to the flagella increasing their diameter. A good species for showing peritrichous flagella (covering the entire cell) is Proteus vulgaris. Some bacteria can protect themselves by secreting large amounts of mucilage around themselves and thereby preventing desiccation or even attack from predators.
The economic importance of bacteria derives from the fact that bacteria are exploited by humans in a number of beneficial ways. Despite the fact that some bacteria play harmful roles, such as causing disease and spoiling food, the economic importance of bacteria includes both their useful and harmful aspects.
Biotechnology and bacteria
Biotechnology or Industrial microbiology is defined as the application of organisms such as bacteria, fungi and algae to the manufacturing and services industries. These include: Fermentation processes, much as brewing, baking, cheese and butter manufacturing, often Lactobacillus in combination with yeasts and moulds, have been used for thousands of years in the preparation of fermented foods such as cheese, pickles, soy sauce, sauerkraut, vinegar, wine and yogurt.
In the chemical industry, bacteria are most important in the production of enantiomerically pure chemicals for use as pharmaceuticals or agrochemicals, such as antibiotics, vaccines and steroids.
Role of the biotechnology industry
200 plus vaccinations products, therapies have been created by the biotechnology industry to combat cancer, autoimmune syndromes, diabetes, HIV/AIDS etc.
Biotechnology industry has developed several environment friendly products treating elements which pollute the environment. Also used in industrial houses for the optimum usage of the various forms of energy. Biotechnology industry also plays a significant role in taking forensic science ahead by using methods relating to DNA fingerprinting.
Geomicrobial prospecting for hydrocarbon exploration
The principle that permits the utilization of microorganisms as sensitive indicators for direct surface sensing of migratory hydrocarbon gases associated with the prospective oil & gas pools appeared meaningful to Mogilveski way back in 1940. He reported that out of 20 microbial anomalies, drilling proved 16. The geomicrobial approach can be used for prospecting stratigraphic traps and can work as an additional tool.
The microbiological survey was first proposed and applied in U.S.S.R. Early use of bacterial soil flora as a means of detecting gas seepages was a development stemming from oil gas surveys performed in U.S.S.R. [1-3]. The initial microbial investigation of Mogilevski and his associates in the field of petroleum prospecting incited the interest of petroleum geochemists worldwide. As a result, microbial methods for detecting petroleum gas in the soils and water have been tested in Europe, USA and elsewhere.
A sustained effort for the purpose of efficiently utilizing microbiological prospecting methods has been underway since 1938 including independent surveys, surveys coordinated with geochemical and geophysical operation, microbiological tests of both soils and subsurface waters and analysis for methane oxidizing as well as higher hydrocarbon oxidizing bacteria. The field trials have been conducted by Sealey [4] in USA, using microbiological techniques. It was shown that of the 89 locations tested in west Texas, wild casts predicted as productive and nonproductive correlation of 54% and 92% respectively. In other series of 42 rank wild cat locations in west central and west Texas, Sealy reported positive prognosis for producers, 8 to 11 and for dry holes 28 out of 31, giving an overall correct prognosis of 85.7%. Miller [5-7] reported microbial survey of a number of oil field in USA, some of which are primarily stratigraphic and showed that microbial activity profiles indicated a good contrast between oil fields and nearby dry areas.In a microbiological and geochemical survey carried out in Hungary, Sozolonki [8], observed strong positive correlation of data with seismic profile and results of drilling.
Boruah [8,9], obtained validity of microbial method for petroleum prospecting through extensive survey of oil fields of Digboi & Naharkatia and a non-petroliferous area in lower Assam Valley, with more than 88% correct prognosis, using this method, they reported a 70% correct prognosis of a productive region Bozaloni and virgin area Jhanji. Some geochemical parameters such as acid soluble minerals, organic carbon and sand were correlated with the geomicrobial data.
After a full of decade, when petroleum discoveries was relatively easy, the attention was again in focus for deploying unconventional methods of surveys for hydrocarbon exploration. Every oil company is carrying out such surveys including geomicrobial survey for risk reduction and better success ratio.
Invisible micro seepages are proof of effective vertical hydrocarbon migration [10,11]. Initially methane and later ethane, propane and butane oxidizing bacteria were exploited for petroleum exploration [12-14] showed that microbial activity profiles in soil cover indicate a good contrast between oil producing and non-producing areas.
Beghtel [15] predicted potential of 18 wildcat wells in Kansas and 13 were proved commercial producer of oil and gas. The success rate was 72%. The integration of microbial survey data with geophysical and other techniques in Yacimientos Petroliferous Fiscalse Bolivianos has proved very successful [16].
Indian oil and gas exploration company, ONGC is carrying out geomicrobial surveys in different basins of India regularly since 1989 and the results have been integrated with geological and geophysical surveys for prioritization of high-risk prospects [17-20].
Growth yields on gaseous hydrocarbons
The growth rates for bacteria on gaseous hydrocarbons can be somewhat long, generation time can be from 4-24 hrs, but the yield of cells per gram or substrate utilized can be rather high. Perry [21] reviewed the topic and presented a table of cell yields from various gaseous hydrocarbon sources (Table 2).
| Substrate | Cell yield (g cells/g substrate) | |
|---|---|---|
| M. vaccaea | P. butanovorab | |
| Ethane | 1.22 | 0.80 |
| Propane | 1.24 | 0.82 |
| n-Butane | 0.72 | 0.83 |
| Ethanol | 0.58 | 0.57 |
| Acetate | 0.18 | 0.16 |
| n-Propanol | 0.70 | 0.95 |
| Acetone | 0.59 | NRc |
| Propionate | 0.45 | 0.75 |
| n-Butanol | 0.46 | 0.85 |
| Butyrate | 0.48 | 0.80 |
Table 2: Cell yields of Mycobacterium vaccae strainJOB5 and Pseudomonas butanovaora grown on various gaseous hydrocarbons and related substrate.
Following table gives an indication of cell yields of M. vaccae strain JOB5 and Pseudomonas butanovora grown on various gaseous hydrocarbons and related substrates (Table 2).
Methane oxidizing bacteria
The principle advantage in using methane oxidizing bacteria for petroleum prospecting is the preponderance of methane in petroleum and it is most mobile of petroleum hydrocarbon predominance of methane in petroleum gas seepage is thus unquestioned. The principle disadvantage in employing methaneoxidizing bacteria as indices of petroleum gas is the relative non specificity of methane to petroleum compared with other hydrocarbon gases such as butane and propane. The later gases have a high degree of specificity with regard to petroleum. Only a few part per million of ethane and propane have been detected in gas over the anaerobic decomposition (fermentation) of the biological production of these gases is not known. In contrast methane is a common product of the anaerobic decomposition of organic matter in nature. It is expected that microorganisms with the ability to utilize the methane are equally widespread in nature. Even though the formation of methane in soil may be intermittent, bacteria are prevalent in soil, which have the capacity to utilize methane when it is made available to them. Methane measured in soil at relatively shallow depth of a few feet is in order of 5.15 ppm in areas that are not associated with petroleum [22-24]. In many areas overlying petroleum accumulation where the methane of soil possible would be a result of petroleum gas-seepage, methane values are not high but show only two fold increase in concentration over so called background (non-petroliferous) areas.
This relatively small contrast in methane concentration is petro bearing areas as compared with non-petroleum bearing areas and the non-specific nature of methane limit its value in petroleum prospecting. The majority of methane oxidizing bacteria do not utilize carbohydrate and proteinaceous culture media. Slavonia reported that methane-oxidizing bacteria grown on peptone media lose their abilities to produce peroxidise required for methane oxidation.
Propane oxidizing bacteria
Bokova isolated propane oxidizing bacteria and identified them as Mycobacterium rubrum. Var propanicum. These produce coral coloured colonies on mineral salt washed agar medium under propane and air. The bacteria do not utilize either methane or ethane but utilize propane and higher paraffin hydrocarbon. These bacteria referred to as Mycobacterium rubrum. Var propanicum grow readily in peptone broth, hydrolyse starch and fats. The bacteria produce visible growth in the presence of propane in media. Utilization of a variety of non-hydrocarbon substrate by propane oxidizing bacteria has been of particular concern regarding their use in petroleum prospecting. Ethane oxidizing bacteria have been used in USSR to a lesser extent due to their infrequent occurrence in some cases of reported biogenied production.

It is of particular interest that propane oxidizing bacteria are obvious choice of soviet scientists in research of petroleum. Brova states that tens of thousands of soil samples taken at depth of 3 m were tested for propane oxidizing microorganism’s microscopic examination of the bacterial pellicle which characteristically form on the supernatant of the mineral salts culture medium usually reveals the presence of bacteria which resemble Mycobacteria propanicum. Not only are propaneoxidizing bacteria of more value than methane oxidizing bacteria of petroleum but bacteriological analysis of spring water and ground water are emphasized more than soil analysis at times. Dostalek has reported isolating propane oxidizing bacteria Pseudomonas from soil of an oil-bearing area Chrechoslovakia. These he refers as Pseudomonas propanica. These bacteria did not utilize methane and ethane but utilize higher hydrocarbon including both paraffinic and cyclic. These Grams negative, both spore forming motile bacteria, utilizes a variety of nonhydrocarbon substrates.
Butane oxidizing bacteria
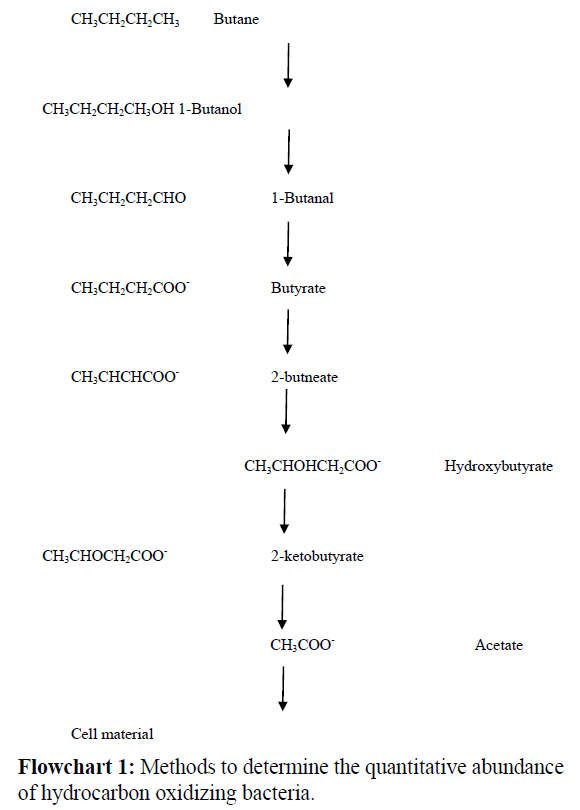
Initial studies on the pathway of metabolism of butane by M. Vaccae strainJOB5 [23] indicated that it is metabolized via acetate which induced isocitrate lyase. This work was later confirmed and extended by Phillips and Perry in 1974 that measured isocitrate lyase, to indicate C2 pathway and 14CO2 incorporation into accumulated pyruvate, to indicate the propionate-methylmalonate pathway, in strainJOB5. Fatty acid profiles were also done to indicate which pathways were present in cells grown on a variety of C4 intermediates. It was discovered that butane was metabolized via acetate and probably through butyrate and β-hydro butyrate with cleavage to form two acetate molecules, which was the usual mode for β-hydroxybutyrate metabolism. 2-butanone apparently underwent a C3+C1 cleavage and was metabolized via the propionate-methyl malonate pathway.
Various methods have been used by different workers to determine the quantitative abundance of hydrocarbon oxidizing bacteria. Some of the common methods used in petroleum prospecting are listed below (Flowchart 1);
• Bacterial pellicle formation
• Gas uptake
• Soil dilution & plating
• Sprinkled soil plating
• Clumped soil planting
• Radio autography
Methodology
Sample collection
Samples for this study were collected by ONGC scientists and were made available to me. Sampling for geomicrobial survey has been carried out on a pre-defined grid pattern of 400×400m. Samples were collected from a depth of about 1 m by digging pits from Natanpur, Hilara (Assam) and collected in presterilized whirl pack bags under aseptic conditions. The samples were stored in cool conditions to arrest the bacterial activities till analysis. A total of 70 sub soil samples were taken up for the study.
Preparation of water blanks
9 ml. of demineralised/distilled water is taken in a test tube. Cotton plugged it and covered with aluminium foil. It was sterilized in an autoclave at 15 psi pressure at 121°C for 15 minutes.
Preparation of stock solution
(i) Potassium nitrate solution- 10 g of Potassium nitrate is dissolved in water to form 100 ml of solution. Sterilize by filtering through 0.22 μm filter.
(ii) Magnesium sulphate solution- 10 g of Magnesium sulphate is dissolved in water to form 100 ml of solution. Sterilize by filtering through 0.22 μm filter.
(iii) Calcium chloride solution- 1.0 g of Calcium chloride is dissolved in water to form 100 ml of solution. Sterilize by filtering through 0.22 μm filter.
Preparation of trace element solution
Following compounds are added sequentially in water to form 1000 ml of solution.
Aluminum hydroxide: 0.10 g
Stannous chloride: 0.05 g
Potassium iodide: 0.05 g
Lithium chloride: 0.05 g
Magnesium Sulphate: 0.08 g
Metaboric acid: 0.50g
Zinc sulphate: 0.10g
Nickel sulphate: 0.05 g
Barium chloride: 0.05 g
Ammonium molybedate: 0.05 g
Preparation of Phosphate buffer
Solution A- 100 ml 0.6 M Sodium hydrogen phosphates.
Solution B- 70 ml 0.6 M Potassium dihydrogen phosphate.
Mix A and B Solution- The final pH value should be 6.8.
Preparation of Agarose base
1.5 g of agarose is dissolved in 100 ml of distilled water in a conical flask. The flask is cotton plugged and covered with aluminium foil. The agarose base is sterilized in an autoclave at 15 psi pressure at 121°C for 15 minutes.
Preparation of mineral media agarose plate
Mineral media of the following composition is prepared by dissolving the salts sequentially in a separately sterilized agarose base at about 50°C. For preparation of 300 ml of medium. Following constituents are added sequentially.
Potassium nitrate: 3ml
Magnesium sulphate: 3ml
Calcium chloride: 3ml
Ferrous sulphate: 0.15 g
Trace element solution: 1 ml
Phosphate buffer: 6 ml
The mineral media Agarose base is poured in pre sterilized disposable Petri plates. The plates are allowed to cool to solidify.
Sterilization of media and apparatus
Agarose base and 9 ml water blanks test tubes are sterilized in an autoclave at 121°C and 15 psi for 15 minutes. Mineral Salt solution and trace element solution are sterilized through Millipore filter paper.
Bacteriological pipettes and spreader are sterilized in electrical oven at 180°C for 2 hrs, after wrapping with aluminium foils.
Disposable ware used
Petri plates
Tips of micropipettes
Equipment’s used
Autoclave
Colony counter
Electronic balance
Laminar airflow
Refrigerator
Deep refrigerator
B.O.D. incubator
Vortex Stirrer
Gas burner
Glassware used
Conical flask
Culture tubes
Spreader
Test tubes
Micropipettes
Beakers
Millipore filter
Measuring cylinder
Procedure
1. Mark test tubes containing 9 ml autoclaved distilled water with sample numbers in sets of 4.
2. Weigh 1g of each soil sample and add in respective test tubes.
3. Inside laminar airflow vortex each test tube for proper mixing of soil in water.
4. Prepare 10-3 and 10-4 dilutions of each sample by serial dilution.
5. Mark the pre-prepared petri plates containing Propanol and Butanol with sample number, dilution factor and date of inoculation. Also mark 4 plates, 2 each of Propanol and Butanol as control.
6. Now add 1 ml of 10-3 and 10-4 dilutions of each sample on respectively marked Agarose plates of Propanol and Butanol and spread them properly using spreader.
7. Place the inoculated petri plates in B.O.D incubator at 36°C for 3 days and finally the developed bacterial colonies are counted in each plate and results are reported as cfu/g of soil (Colony Forming Units per gram of soil).
Result and Discussion
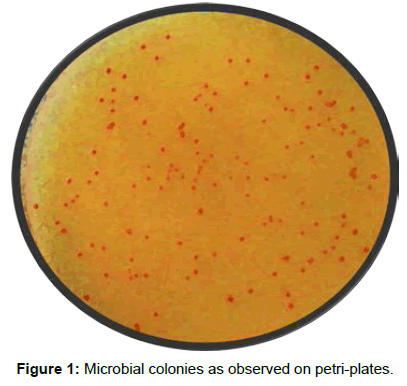
The abundance of propane and butane oxidizers in soil samples was calculated by observing and counting the developed colonies of respective gaseous hydrocarbon oxidizers after an incubation period of 3 days at 37°C (Table 3). The bacterial colonies observed on plates are shown in Figure 1.
| Sample No. | Propane Oxidizers | Butane Oxidizers | ||||
|---|---|---|---|---|---|---|
| Viable Count (x103) | Viable Count (x104) | Viable Count (x103) | Viable Count (x104) | |||
| 381 | 240 | 56 | 252 | 28 | ||
| 382 | 112 | 22 | 140 | 20 | ||
| 400 | 34 | 3 | 56 | 4 | ||
| 402 | NIL | NIL | 6 | 1 | ||
| 404 | 90 | 10 | 60 | 6 | ||
| 406 | 5 | NIL | 25 | 4 | ||
| 411 | 2 | NIL | 40 | 5 | ||
| 413 | 170 | 12 | 720 | 77 | ||
| 414 | 3 | NIL | 90 | 5 | ||
| 421 | 6 | 1 | 53 | 9 | ||
| 422 | NIL | NIL | 7 | 1 | ||
| 424 | 17 | 2 | 45 | 11 | ||
| 429 | 2 | NIL | 8 | NIL | ||
| 430 | 2 | NIL | 9 | 1 | ||
| 431 | 34 | 4 | 8 | 2 | ||
| 435 | 6 | NIL | 18 | 5 | ||
| 437 | 35 | 6 | 141 | 15 | ||
| 438 | 8 | 1 | 16 | 3 | ||
| 439 | 15 | 2 | 42 | 8 | ||
| 440 | 6 | 1 | 8 | 6 | ||
| 443 | 100 | 9 | 72 | 10 | ||
| 444 | 20 | 3 | 19 | 3 | ||
| 445 | 6 | NIL | 8 | 1 | ||
| 446 | 67 | 5 | 112 | 14 | ||
| 447 | 16 | 2 | 50 | 7 | ||
| 448 | 59 | 5 | 7 | NIL | ||
| 450 | 10 | 1 | 21 | 20 | ||
| 451 | 4 | NIL | 17 | 1 | ||
| 452 | 3 | NIL | NIL | NIL | ||
| 453 | 12 | 1 | 11 | 2 | ||
| 454 | 5 | NIL | 10 | 1 | ||
| 455 | 15 | 5 | 20 | 3 | ||
| 456 | 13 | 4 | 27 | 3 | ||
| 457 | 76 | 5 | NIL | NIL | ||
| 460 | 4 | NIL | NIL | NIL | ||
| 461 | 33 | 5 | 16 | 2 | ||
| 462 | 11 | 2 | 22 | 3 | ||
| 463 | 6 | 2 | 23 | 4 | ||
| 464 | 24 | 4 | 32 | 4 | ||
| 465 | 7 | 1 | 20 | 3 | ||
| 466 | 15 | NIL | 13 | 2 | ||
| 467 | 42 | 7 | 9 | 2 | ||
| 471 | 7 | NIL | 40 | 6 | ||
| 472 | 16 | 2 | 43 | 6 | ||
| 473 | NIL | NIL | NIL | NIL | ||
| 474 | NIL | NIL | 2 | NIL | ||
Table 3: Propane and Butane oxidizing bacterial Count in Soil Samples.
All the samples were processed in duplicate and plates showing colonies 30-300 were taken into consideration. Average of two plates (duplicate processing) as well as all the valid dilutions was considered, while the contaminated plates/doubtful results were ignored.
The results of propane and butane oxidizers population will be plotted in terms of population density using SURFER-7 software after evaluation of total samples (500-700 no.) (Table 3).
Conclusion
• It is observed that propane oxidizers count varies from NIL to 56x104 and butane oxidizers count varies from NIL to 77x104.
• The samples showing high values of propane and butane oxidizers bacteria indicate the proximity of hydrocarbon source.
For any geomicrobial survey for hydrocarbon exploration a large number of samples to the tune of 500-1000 are to be analysed and the area encompassing the higher value sample is demarcated as anomalous zone. This bacterial anomalous zone is integrated with other petroleum survey techniques for finalizing an area rich in hydrocarbons for drilling of an oil well (Figure 1).
References
- Dostalek MM. Propane-oxidising bacteria, Gesk. Biol. 1954;3:173-181.
- Chandra D, Biswas D, Mishra AK, et al. Geochemical and Geomicrobial prospecting of oil and gas. 1987.
- Dalton H. Oxidation of hydrocarbons b methane monooxygenases from a variety of microbes, Appl. Microbial. 1980;26:71-87.
- Sealey LO. A Geomicrobiological method of prospecting for petroleum (Part 1 & 2). Oil and Gas Jour. 1974:98.
- Miller. Microbial survey helps to evaluate oil and gas. Oil and gas jour. 1976;4:192.
- Craggs BA, Parkes RJ, Fry JC, et al. Bacterial population and processes in sediments containing gas hydrate. Earth and Planetry Science Letters. 1996;139:497-507.
- Higgins IJ, Best DJ, Hammon RC, et al. Methane Oxidising Microorganism. Microbial Revs. 1981;45:556-590.
- Sozolanki J. Exploration of hydrocarbon deposits by microbiological and direct geochemical methods in the area around Norahalm. 1978.
- Boruah C, Bora BK, Pillae KR, et al. Geomicrobial and Geochemical approaches to Petroleum prospecting. Asia Journal. 1984;-190- 195.
- Colby J, Dalton H, Whittenberg R. Biological and Biochemical aspects of microbial growth on C1 compounds. Rev. Microbial. 1979;33:481-517.
- Colwell FS, Delwiche ME, Black welder D, et al. Microbial communities from core intervals, JAPEX/GSC Malik 2L-38 gas hydrate research well, Geological survey of Canada, Bulletin. 1999;544:189-195.
- Indrani R, Shukla SK, Mishra AK, et al. Microbes as an indicator of underground petroleum deposits. Fuel. 1989;68:311-314.
- Hoffman B, Rehm HJ. In Biotechnology Vol. 6a (ed. H.J.Rehm) VCH, mbH, FRG. 1984.
- Martens CS, Chanton JP, Paull CK. Biogenic methane from abyssal brine seeps at the base of the Florida Eacarpment. Geology. 1991;66:157-168.
- Beghtel F. Microbial oil survey technique (MOST) evaluation of new field wildcat wells Kansas, APGE Bull. 1987;3(1):1-14.
- Pareja L. Combined Microbial, Seismic Survey Predict Oil And Gas occourance in Bolivia ,oil and gas journal. 1994:68-70.
- Jain AK, et al. Geomicrobial survey in Kangra-Paror area in Himalayan foothills of Himachal Pradesh, KDMIPE Report. 2001.
- Brisbane PG, Ladd JN. Role of microorganism in petroleum exploration. Ann Appl Microbial. 1965;26:71-87.
- Claypool GW, Kaplan IR. The origin and distribution of methane in marine sediments. In natural gases in marine sediments eds. Kaplan I.R. 99-139, Plenum Press, New York. 1974.
- Hanson RS. Ecology and Diversity of Methylotrophic organism. Adv. Appl. Microbial. 1980;26: 3-39.
- Perry JJ. Propane utilization by microorganisms. Adv. Applied Microbiology. 1980;26:89-115.
- Kartsev AA, Teals. Geochemical methods of prospecting and exploration for petroleum natural gases, California press, Berkeley California. 1959.
- Vastal JR, Perry JJ. Divergent metabolic pathways for propane and propionate utilization by a soil isolate. J. Bcaterial. 1968;99:216-221.
- Schumacher DD. Surface Geochemical Exploration for oil and gas: new life for an old technology. The leading Edge. 2000:24-26.