Research Article - Biomedical Research (2017) Volume 28, Issue 1
Microbial community structure in saliva from children of the Baoan nationality with or without dental caries in gansu, China
Jianye Zhou1, Yujuan Zhu2, Zhanhai Yu3, Kangli Jiao1, Fang Wu3 and Zhiqiang Li1*1Key Laboratory of Oral Diseases of Gansu Province, Northwest University for Nationalities, Lanzhou PR China
2Tai An, TSCM Hospital, PR China
3School of Stomatology, Lanzhou University, Lanzhou, Gansu, PR China
- *Corresponding Author:
- Zhiqiang Li
Doctor of Science
Key Laboratory of Oral Diseases of Gansu Province
Northwest University for Nationalities
PR China
Accepted date: May 21, 2016
Abstract
Purpose: The purpose of this study was to compare the microbial colony composition in the saliva of children with and without caries in a regional ethnic group (Baoan) in China.
Materials and methods: The recruited participants were aged 5-9 years old and from Bonan district, China. Eighteen samples (10 from subjects with caries and 8 from subjects without caries) were taken and divided into two groups respectively (BC and BH). These samples characterized by 16S ribosomal RNA (16S rRNA) were studied using cloning technology. A total of 900 clones were sequenced and 860 qualified reads were obtained for evaluating bacterial diversity.
Results: The overall operational taxonomic units (OTUs) distribution of 16S rRNA gene clones indicated the difference between dental caries and caries-associated salivary microorganisms. The phyla of the samples with caries were the same as caries-free samples, namely: Firmicutes, Proteobacteria, Actinobacteria, Fusobacteria and Bacteroidetes. The family Micrococcaceae and the genus Rothia showed significant difference (p<0.05) between BC and BH groups, the amount of which were higher in BC group.
Conclusion: Our data demonstrated that, at the phyla level, the predominant phyla in saliva of Baoan Children are relatively constant. However, at the genus level, the composition of microbial colony in dental caries and healthy teeth were of significant difference.
Clinical relevance: The attention on the prevention of caries in children should not be only paid to the traditional cariogenic bacteria, but also to the microbial colony composition of saliva.
Keywords
Dental caries, Microbial diversity, Saliva
Brief Synopsis
The family Micrococcaceae and the genus Rothia showed significant difference in the microbial colony composition of caries compared to healthy teeth.
Abbreviations
OUTs: Operational Taxonomic Units; DMFT: Number of Decayed (D), Missing (M) and Filled (F) Teeth (T); PCoA: Principal Coordinates Analysis.
Introduction
Dental caries is a common oral childhood disease with a high prevalence worldwide. It is also a major problem for children in China [1], which affects chewing, aesthetics, and speaking, thus exerting significant influence on the physical and mental health as well as their life span. Children’s teeth are especially susceptible to caries partly because their immune systems are not yet fully developed and their salivary glands are not completely mature [2-6]. Therefore, it is beneficial to detect the unique cariogenic bacteria in children and take measures to prevent dental caries in childhood. Prevention of dental caries in children is always among the first priorities of national health policies because children’s oral health is an integral part of general health. Therefore, it is a goal of growing importance to establish accurately the characteristics and causes of caries in children. It is widely accepted that the etiology of dental caries includes bacteria, oral environment, host and time [7]. Out of these four factors, bacteria are clearly the most important and many prevention studies have focused on S. mutans, Lactobacillus and S. sobrinus that have always been considered the major pathogenic bacteria [8]. A long-term study showed that dental caries were caused by a combination of a variety of bacteria and not by a singular bacterium [9]. Another investigation demonstrated that the diversity of the microbial colony composition in saliva influenced the development and incidence of caries and other oral diseases [10]. In addition, others demonstrated that the composition of the bacterial colony was much more important to the development of caries than the mere presence of a potentially pathogenic species [11,12].
At the same time, it is becoming more notable that oral health or disease depends on the interface between the host and the microbes. Current studies show that the genetic background of the host can influence the microbial colony composition of the oral cavity [13,14]. Therefore, understanding the host is the key to combating human oral diseases.
The oral bacterial microbiome encompasses approximately 700 commonly occurring phenotypes. With the help of ribosomal RNA-based molecular taxonomy, bacterial colony analysis can be performed to identify the interactions between the various phenotypes, and between the phenotypes and their environment. In addition, bacterial colony compositions are related to disease initiation and disease progression [15]. There are many factors that influence the formation and the evolution of microbes [16]. A better understanding of how these factors affect the composition of the oral microbial colony can help to define a new strategy for preventing oral disease.
In our current study, we chose subjects belonging to a regional ethnic group in China, in order to investigate the structure of oral bacterial colonies for a specific population. Meanwhile, with the identification of the differences between caries and healthy samples, which provide some clues to how bacterial colony composition affect oral disease and how it is monitored by microbial interactions, thus leading to new strategies for management of oral disease.
Methods
Subjects and specimen collection
Eighteen child volunteers were chosen from Bonan autonomous county in Gansu province, China, in September 2013. The eligibility criteria included that the last three generations of the candidates had to be of the same ethnicity. The children were fully screened according to the research requirement and their guardians agreed to sign informed consent, required by the Ethics Committee of Lan Zhou University. The age of the recruited children ranged from 5 to 9 years. A complete clinical examination was performed on all the children, including an intra-oral examination and a fullmouth periodontal probing. According to the recorded medical and dental examination reports, all children were free of systemic diseases, periodontal diseases (periodontitis and gingivitis) during a 24-hour observation period (exclusion criteria). Then all subjects were examined using the dmft (decayed, missing and filled teeth) index. The subjects were then divided into two groups, namely BC (10 Bonan subjects with caries: dmft index ≥ 4) and BH (8 Bonan healthy subjects: dmft index <4) according to dmft index defined by the World Health Organization (Table 1). Their dietary habits (breakfast, lunch, dinner, snacks and soft drinks) were surveyed using a questionnaire and we found no significant difference between the two groups. Their diet mainly consisted of noodles and potatoes and nearly no snacks or soft drinks. Clinical examination was performed by a single trained examiner in order to ensure consistency of clinical data.
| Sample ID | Gender | Age | Ethnic | DMFT index |
| BH027 | M | 5 | Bonan | 0 |
| BH069 | M | 8 | Bonan | 0 |
| BH104 | F | 8 | Bonan | 0 |
| BH110 | M | 6 | Bonan | 0 |
| BH409 | M | 9 | Bonan | 0 |
| BH410 | F | 9 | Bonan | 0 |
| BH413 | F | 8 | Bonan | 0 |
| BH444 | F | 9 | Bonan | 0 |
| BC103 | F | 5 | Bonan | 4 |
| BC106 | M | 9 | Bonan | 4 |
| BC115 | F | 8 | Bonan | 4 |
| BC182 | F | 6 | Bonan | 5 |
| BC187 | M | 7 | Bonan | 6 |
| BC191 | M | 6 | Bonan | 5 |
| BC334 | M | 8 | Bonan | 6 |
| BC376 | M | 9 | Bonan | 4 |
| BC380 | F | 7 | Bonan | 7 |
| BC431 | M | 8 | Bonan | 6 |
Table 1: Characteristics of subjects and DMFT index in this study.
DNA extraction
One-milliliter of unstimulated saliva samples was collected from each subject using an aseptic tube containing 500 μL TE buffer (25 mM Tris-HCl, 10 mM EDTA, pH 8.0). Saliva was only collected at one time point, because it can be assumed that the bacterial composition of saliva does not change over a 6-h time interval. The samples were taken to the laboratory immediately after the clinical parameters were recorded. Ten microliters of lysozyme (100 mg/mL) was added to each sample, and incubated at 37ºC for 24 hours in a shaker (200 r/ min). Following this the samples were incubated at 65ºC for 1 hour in proteinase K (20 mg/mL) and boiled for 10 minutes after the addition of 180 μL of NaOH (50 mM). Subsequently, the DNA was crude-extracted using trichloromethane (1:1) and precipitated with 70% ethanol and re-suspended with an appropriate amount of TE buffer [17].
PCR amplification and product purification
The 16S rRNA region, which is approximately 1.5 Kbp was amplified with primers 8 F (5’ - AGAGTTTGATCCTGGCTCA - 3’) and 1492 R (5’ - GGTTACCTTGTTACGACTT - 3’) in a final reaction volume of 50 μL (2 μL of 10 mM dNTPs, 1 μL of each primer, 0.5 μL of Taq DNA polymerase Takara, 10× buffer). The amplification cycle was initiated by a process of denaturation at 94ºC for 5 minutes, followed by 30 cycles of template denaturation at 94ºC for 40 seconds, then annealing at 54ºC for 45 seconds, and extension at 72ºC for 1.5 minutes, lastly a final extension at 72ºC for 10 minutes. The resultant PCR products were confirmed by 1% agarose gel electrophoresis and purified using a QIA-quick PCR purification kit (Takara, Dalian, China).
16S rRNA gene clone and sequencing
Purified amplicons were ligated into plasmid vector PMD18-T (Takara, Dalian, China) and transformed into Escherichia coli using the Takara Cloning kit (Takara, Dalian, China) according to the manufacturer’s instructions. The correct insert sizes were determined by PCR using M13 forward and reverse primers [18,19]. The size of the inserts (approximately 1.5 Kbp) was determined by PCR with flanking vector primers followed by electrophoresis on a 1% agarose gel. Single-track sequencing was performed on the plasmid inserts of the purified PCR products (Life Genomics Institute, Beijing, China).
Statistical analysis
Statistical analysis involved software SPSS 13.0, with ANOVA or Student’s t-test being applied as appropriate. p<0.05 was deemed significant, and p<0.01 highly significant.
Results
Information of the sequencing and diversity index
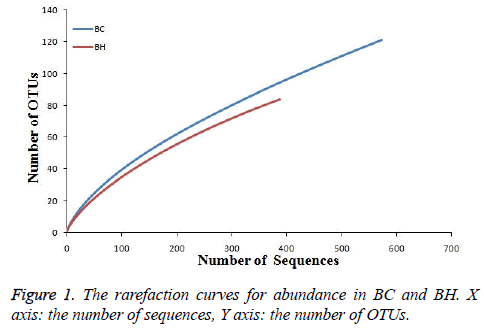
The vector sequence regions were removed after processing with the software VecScreen, and low-quality sequences with chimeras were removed. The sequences were aligned and edited using ClustalW and BioEdit software, respectively. Operational taxonomic units (OTUs; phenotypes) were defined using a 97% sequence similarity cutoff, which roughly corresponds to species-level groupings. One representative sequence from each OTU identified in this study was deposited in the RDP database (http://rdp.cme.msu.edu/). The 900 sequences (BC: 500, BH: 400) that were analyzed using Mothur software with a cutoff of 97% were grouped into 170 OTUs. These OTUs belonged to 5 phyla, 12 classes, 16 orders, 23 families, and 28 genera. The BC group covered 121 OTUs while the BH group covered 84 OTUs (there were 35 OTUs in common between the two groups). The coverage of the two libraries was 0.86 (BC) and 0.87 (BH) using the coverage formula (1 - (n / N)) × 100 (n is the number of singleton phenotypes, and N is the total number of clones). The microbial diversity of both groups was similar. There was no significant difference between the two libraries in either the Shannon Index or the Simpson Index (p>0.05), which was calculated by T-test.
Colony structure and variance
The rarefaction curves for the abundance of BC and BH were made to evaluate the reliability of our data (Figure 1). The curve tended to smoothness with the increasing number of sequences, suggesting the reliability of our data.
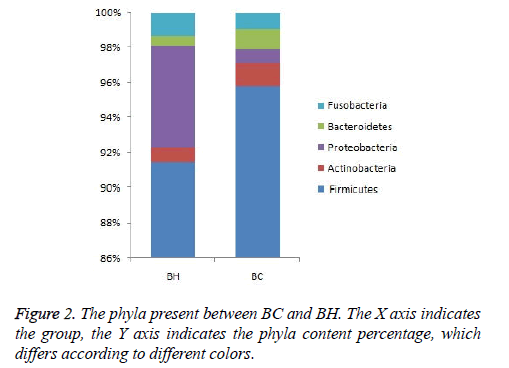
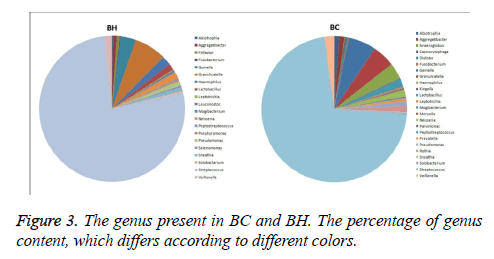
The same kind of OTUs were clustered at the level of phyla and genus, and using the Mothur software (http:// www.mothur.org/), significant differences between libraries were found by T-test. The phyla in BC and BH were the same: Firmicutes, Proteobacteria, Actinobacteria, Fusobacteria, and Bacteroidetes (Figure 2). The dominant genus in the two libraries is listed in Figure 3. The dominant genera in BC were: Streptococcus (71.8%), Gemella (6.2%), Granulicatella (5%), Haemophilus (3%), Veillonella (2.2%), Lactobacillus (1.7%), Neisseria (1.5%), Rothia (1.33%), Aggregatibacter (1.2%), Abiotrophia (1.1%), and Pseudomonas (1%). While the dominant genera in BH were: Streptococcus (77%), Granulicatella (7.2%), Gemella (3.4%), Haemophilus (2.1%), Veillonella (1.6%), Neisseria (1.5%), Lactobacillus (1.4 %) and Pseudomonas (1.1%). The family Micrococcaceae and the genus Rothia showed significant difference (p<0.05) between BC and BH groups, being higher in the BC group, while the family Peptostreptococcaceae was higher in the BH group.
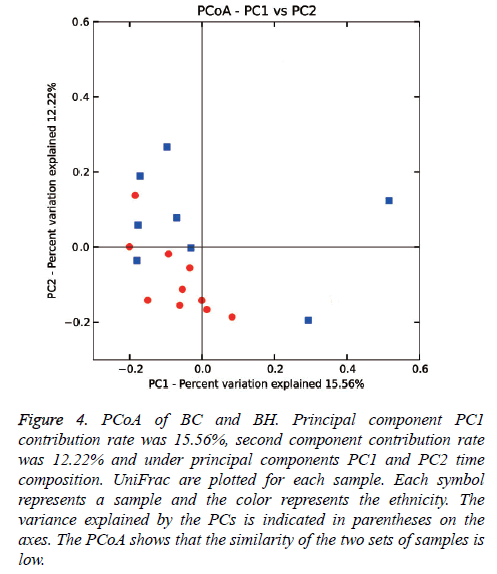
Weighed and normalized PCoA was performed to evaluate similarity among samples, with each sample representing an environment. Based upon the 2 primary vectors, the significance was evaluated using a two-tailed T-test (Figure 4). The PCoA data showed that there was a difference between the microbial colony structures of the two groups.
Figure 4: PCoA of BC and BH. Principal component PC1 contribution rate was 15.56%, second component contribution rate was 12.22% and under principal components PC1 and PC2 time composition. UniFrac are plotted for each sample. Each symbol represents a sample and the color represents the ethnicity. The variance explained by the PCs is indicated in parentheses on the axes. The PCoA shows that the similarity of the two sets of samples is low.
Discussion
Bonan are an ethnic minority that live in the shadow of Mountain Jishi near the Yellow River in the Gansu province of China. Bonan population was nearly 1.6 million in this region, which constitutes 80% of this ethnicity in China. Due to their geographical remoteness, inaccessibility and poor economy, the daily diet comprises of pasta and potatoes. A description of this population’s oral microbe composition may provide important information pertaining to the oral microbial colony composition of children with caries. As stated in the introduction, dental caries is caused by a combined effect of a variety of bacteria and not by a singular bacterium. Plenty of evidence has shown that the presence of microbial colonies is important in the development of oral caries.
Saliva has been considered to be the most suitable sample to provide information related to the microbial colony composition of the entire oral cavity due to its universality and accessibility. On one hand, saliva contains a variety of bacterial species from different areas of the mouth (e.g., tongue, subgingival plaque, and supragingival plaque) [20,21]. A mixture of the microbial consortia that existed at various sites in the oral cavity and the microbial populations in saliva are relatively stable over time. On the other hand, saliva is an easily and inexpensively accessible biological fluid, and it has been thoroughly analyzed for biomarkers of health and disease [22-25]. According to previous studies, 16S rRNA clone library technology with specific primers can be applied to obtain 16S rRNA gene fragments from these saliva samples. The achieved PCR products are cloned into a plasmid vector and then sequenced and compared with the 16S rRNA sequences extant in databases. Therefore, the 16S rRNA clone library method has been considered optimal and applicable for analyzing the dominant species in the oral microbial pool for healthy patients [26] or those with oral diseases [27-30]. As stated above, 16S rRNA clone library continues to be regarded as a promising method due to the fact that it can save the plasmids and strains for subsequent studies and offers more accurate sequencing and longer reads. All of these advantages enabled us to study the microbial colony composition in saliva of children with and without caries.
Our study focused on a regional ethnic group of Baoan in China, and exclusively explored the oral bacterial colonies. Though the amount of the samples was limited, we ruled out the ethnic diversity in the comparison of the microbial colony composition between caries-active and healthy oral cavities. At the phyla level, the phyla of microbial coolony in the caries samples were the same as in the caries-free samples (Firmicutes, Proteobacteria, Actinobacteria, Fusobacteria and Bacteroidetes), though they were in different proportions. These phyla are the most common phyla as reported in previous studies on children’s caries [31-35] and simultaneously, these phyla are largely comparable to the most common phyla in adults [36]. It is indicated that the predominant phyla in saliva is relatively constant, though the proportion varies by ethnicity and genetic makeup.
However at the genus level, the PCoA showed that there was a difference between the microbial colony compositions in caries compared to healthy individuals. The family Micrococcaceae and the genus Rothia were higher in caries group, while the family Peptostreptococcaceae was higher in the healthy group. Rothia belongs to the family Micrococcaceae and is an aerobic or micro-aerobic Gram-positive bacterium. The true role of Rothia in caries is not clear yet. One study showed that it was an opportunistic pathogen in adults [37] while another study showed that it was one of the most predominant bacterial genus in the oral cavity [38]. In view of the abundant diversity of microbial colony composition in saliva, there was no significant difference at the genetic level, which is believed to maintain internal metabolic stability. Certain caries-active microbial colonies such as Rothia may participate in complex metabolism of carbohydrates, nitrogen and amino acids, involved in the development of caries. In which case, further validation is needed for early prevention of dental decay. Children are highly susceptible to caries as the cariogenic bacteria are more active than that in adults and it is interesting to find that Streptococcus and Lactobacillus, which are associated with adults caries, do not seem to play a significant role in children. Our result on Baoan children indicates that dental caries may be caused by a joint effect of certain oral bacteria and their metabolic by products. Extensive research with large amount of samples involving diverse ethnic population is required to compare and validate current findings in the future. However, the microbial colony composition in saliva should be considered for the prevention of the caries in children.
Availability of supporting data
The data sets supporting the results of this article are available in the GenBank (National Center for Biotechnology Information) repository, [unique persistent identifier and hyperlink to dataset in https://submit.ncbi.nlm.nih.gov/subs/ genbank/].
Acknowledgements
We thank all the volunteers who agreed to participate in this study, the State Ethnic Affairs Commission Key Laboratory of Oral Medicine and the Shanghai parson's biological technology co., LTD (Shanghai, China). These works are financially supported by the and area fund projects of the Chinese National Natural Science Fund (31360124/C0309) and Gansu province science and technology plan (144WCGA167).
References
- Zhang S, Lo ECM, Liu J, Chu CH. A review of the dental caries status of ethnic minority children in China. J immigr minor health 2013; 17(1): 1-13.
- AbdelAziz WE, Dowidar KM, El Tantawi MM. Association of Healthy Eating, Juice Consumption, and Bacterial Counts with Early Childhood Caries. Pediatric dentistry 2015; 37: 462-467.
- Li Y, Ku C, Xu J, Saxena D, Caufield P. Survey of oral microbial diversity using PCR-based denaturing gradient gel electrophoresis. J dent res 2005; 84: 559-564.
- Kanasi E, Johansson I, Lu SC, Kressin NR, Nunn ME, Kent R. Microbial risk markers for childhood caries in pediatricians’ offices. J dent res 2010; 89: 378-383.
- Soncini JA, Kanasi E, Lu SC, Nunn ME, Henshaw MM, Tanner AC. Oral microbiota of children in a school-based dental clinic. Anaerobe 2010; 16: 278-282.
- Gambhir RS, Singh S, Singh G, Singh R, Nanda T, Kakar H. Vaccine against Dental Caries-An Urgent Need. J Vaccines Vaccin 2012; 3.
- Kidd E. The implications of the new paradigm of dental caries. J Dent 2011; 39: S3-S8.
- Takahashi N, Nyvad B. The Role of Bacteria in the Caries Process Ecological Perspectives. J Dent Res 2011; 90: 294-303.
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH. The human oral microbiome. J Bacteriol 2010; 192: 5002-5017.
- Chang RCC, So KF. Use of anti-aging herbal medicine, Lycium barbarum, against aging-associated diseases. What do we know so far? Cell Mol Neurobiol 2008; 28: 643-652.
- Gross EL, Beall CJ, Kutsch SR, Firestone ND, Leys EJ, Griffen AL. Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. Plos One 2012; 7: e47722.
- Peterson SN, Snesrud E, Liu J, Ong AC, Kilian M, Schork NJ. The dental plaque microbiome in health and disease. Plos One 2013; 8: e58487.
- Yang F, Zeng X, Ning K, Liu KL, Lo CC, Wang W. Saliva microbiomes distinguish caries-active from healthy human populations. ISME J 2011; 6: 1-10.
- Mason MR, Nagaraja HN, Camerlengo T, Joshi V, Kumar PS. Deep sequencing identifies ethnicity-specific bacterial signatures in the oral microbiome. Plos One 2013; 8: e77287.
- Robert J, Palmer JR. Composition and development of oral bacterial communities. Periodontol 2000 2014; 64: 20-39.
- Xu H, Hao W, Zhou Q, Wang W, Xia Z, Liu C. Plaque bacterial microbiome diversity in children younger than 30 months with or without caries prior to eruption of second primary molars. Plos One 2014; 9: e89269.
- Noorda W, Purdell-Lewis D, van Montfort A, Weerkamp A. Monobacterial and mixed bacterial plaques of Streptococcus mutans and Veillonella alcalescens in an artificial mouth: development, metabolism, and effect on human dental enamel. Caries Res 1988; 22: 342-347.
- Bradshaw D, Marsh P. Analysis of pH–driven disruption of oral microbial communities in vitro. Caries Res 1998; 32: 456-462.
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. Plos Biol 2008; 6: e280.
- Takeshita T, Nakano Y, Kumagai T, Yasui M, Kamio N, Shibata Y. The ecological proportion of indigenous bacterial populations in saliva is correlated with oral health status. ISME J 2008; 3: 65-78.
- Belstrøm D, Fiehn NE, Nielsen CH, Kirkby N, Twetman S, Klepac‐Ceraj V. Differences in bacterial saliva profile between periodontitis patients and a control cohort. J Clin Periodontol 2014; 41: 104-112.
- Giannobile W, McDevitt J, Niedbala R, Malamud D. Translational and clinical applications of salivary diagnostics. Adv Dent Res 2011; 23: 375-380.
- Farnaud SJ, Kosti O, Getting SJ, Renshaw D. Saliva: physiology and diagnostic potential in health and disease. Sci World J 2010; 10: 434-456.
- Greabu M, Battino M, Mohora M, Totan A, Didilescu A, Spinu T. Saliva-a diagnostic window to the body, both in health and in disease. J Med Life 2009; 2: 124-132.
- Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J 2010; 4: 962-974.
- Schirrmeister JF, Liebenow AL, Pelz K, Wittmer A, Serr A, Hellwig E. New bacterial compositions in root-filled teeth with periradicular lesions. J Endodont 2009; 35: 169-174.
- Subramanian K, Mickel AK. Molecular analysis of persistent periradicular lesions and root ends reveals a diverse microbial profile. J Endodont 2009; 35: 950-957.
- Faveri M, Gonçalves L, Feres M, Figueiredo L, Gouveia L, Shibli J. Prevalence and microbiological diversity of Archaea in peri‐implantitis subjects by 16S ribosomal RNA clonal analysis. J Periodontal Res 2011; 46: 338-344.
- Zhang C, Hou B, Zhao H, Sun Z. Microbial diversity in failed endodontic root-filled teeth. Chin Med J (ENGL) 2012; 125: 1163-8.
- Crielaard W, Zaura E, Schuller AA, Huse SM, Montijn RC, Keijser BJ. Exploring the oral microbiota of children at various developmental stages of their dentition in the relation to their oral health. BMC Med Genomics 2011; 4: 22.
- Luo A, Yang D, Xin B, Paster B, Qin J. Microbial profiles in saliva from children with and without caries in mixed dentition. Oral Dis 2012; 18: 595-601.
- Jiang W, Zhang J, Chen H. Pyrosequencing analysis of oral microbiota in children with severe early childhood dental caries. Curr Microbiol 2013; 67: 537-542.
- Xin BC, Luo AH, Qin J, Paster BJ, Xu YL, Li YL. Microbial diversity in the oral cavity of healthy Chinese Han children. Oral Dis 2013; 19: 401-5.
- Nasidze I, Li J, Schroeder R, Creasey JL, Li M, Stoneking M. High diversity of the saliva microbiome in Batwa Pygmies. PloS one 2011; 6: e23352.
- Rafi W, Venkataswamy MM, Ravi V, Chandramuki A. Rapid diagnosis of tuberculous meningitis: A comparative evaluation of in-house PCR assays involving three mycobacterial DNA sequences, IS6110, MPB-64 and 65 kDa antigen. J Neurol Sci 2007; 252: 163-168.
- Ghoshal U, Kishore J, Kumar B, Ayyagari A. Serodiagnosis of smear and culture-negative neurotuberculosis with enzyme linked immunosorbent assay for anti A-60 immunoglobulins. Indian J Pathol Micr 2003; 46: 530-534.
- Bera S, Shende N, Kumar S, Harinath B. Detection of antigen and antibody in childhood tuberculous meningitis. Indian J Pediatr 2006; 73: 675-679.
- Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol 2005; 43: 5721-5732.