Research Article - Journal of Biochemistry and Biotechnology (2018) Volume 1, Issue 1
Method enabling detection of single-stranded DNA ligation activity.
Tommy Nordström and Pål Nyrén*
School of Biotechnology, Royal Institute of Technology, Roslagstullsbacken 21 SE-114 17 Stockholm, Sweden
- *Corresponding Author:
- Pål Nyrén
School of Biotechnology
The Royal Institute of Technology
Roslagstullsbacken 21
SE -11417 Stockholm, Sweden
Tel: +46 703971630
E-mail: nyren@kth.se
Accepted date: November 29, 2017
Abstract
The enzyme DNA ligase is an essential tool in many different applications in the field of biotechnology. For these applications the DNA quality can be crucial for the final result. In this paper we present a new method that can be used for specific studies of DNA ligases and for simultaneous analyzes of DNA quality. More specifically, a simple, fast and sensitive method for detection of single-stranded DNA (ssDNA) ligase activity is presented. The new method is based on real-time detection of nucleotide incorporation over the ligation site catalyzed by a DNA polymerase. The ratio between the incorporation signal after and before the ligation site gives an estimate of the ligation efficiency. To further increase the robustness of the assay the exact amount of unligated product can be easily estimated in a separate assay using a 3’-end blocked oligonucleotide functioning as template. Produced circular DNA can be cleaned from unligated DNA by exonuclease I treatment and the quality of the circle can be easily analyzed by a simple sequencing procedure. If the ssDNA used for the ligation reaction contains truncated fragments this will be observed by uneven signals during the analysis. The new method can be a useful tool for researchers working with both basic and applied projects in the field of biotechnology.
Keywords
Pyrosequencing, DNA sequencing, DNA ligase, Luciferase, Single-Stranded DNA, Circular single-Stranded DNA, Alkaline phosphatase, Exonuclease I, Inorganic pyrophosphatase
Introduction
Enzymes are important biocatalyst involved in nearly all metabolic processes in living cells. They are of great interest for both basic and applied research in many different fields. During the last decades enzymes involved in DNA metabolism have been in focus and attracted strong interest due to several breakthroughs in the DNA technology field. One group of enzymes of importance for several new inventions in the DNA field is DNA ligases. DNA ligases are essential constituents of all known organisms due to their important roles in the replication, recombination, and repair of DNA [1-3]. They catalyze the formation of a phosphodiester bond between adjacent 5’-phosphoryl and 3’-hydroxyl termini at single-strand nicks in double-stranded DNA [4]. Two types of DNA ligase have been identified that are categorized by whether NAD+ or ATP is used as the source of adenylate that is used to activate the enzyme.
DNA ligases are regarded as valuable molecular tools and can be used for a variety of applications in molecular biology, e.g. in the detection of specific nucleic acid sequences (DNA or RNA) or protein analytes [5-7], in the ligase chain reaction for detection of mutations [8,9] and polymorphisms [10], and in the generation of sequencing primers by random ligation of hexamers [11,12].
Several methods have been developed for analysis of DNA ligation activity. Earlier methods were based on detection of acid-insoluble phosphatase-resistant radioactive-labeled DNA [13] and detection of exonuclease III resistant circulated singlestranded DNA [14]. The most common methods used for analyzing ligase activity are based on electrophoresis in agarose or polyacrylamide gels and detection by autoradiography or staining [4,15,16]. Besides electrophoretic techniques commonly used for detection of ligation activity, several new approaches have been described. Examples for new approaches are methods based on the molecular beacon concept [17] and on electrochemical technique [18].
Many of the methods have limitations in sensitivity, specificity and speed. In this paper we present a new strategy for faster and simpler analysis of DNA ligation activity. The method has been used for analysis of single-stranded DNA circulization but can in principle be used for analysis of double-stranded DNA ligation.
Materials and Methods
Synthesis and purification of oligonucleotides
The oligonucleotide 80 mer Probe (5’-Phos-GCGTCA GCGGGTGTTGGCGGGTGTCGGGGCTGGCTTAAC TATGCGGCATCAGAGCAGATTGTACTGAGAGTG CACCATAT), primers for detection of ligation activity Seqv-1Lsite (5’CCGCCAACACCCGCTGACG) and Puc_Seqv.CL113 (5’CACCCGCCAACACCCGCT), oligonucleotide for detection of unligated product Ex non LP (5’-AGGAGATATGGTGCACTCTCAGT-Phos), and PCR primers Puc_UP125 (5’-Phos-GCGTCAGCGGGTGTTG) and Puc_Do168 (5’-Biotin-ATATGGTGCACTCTCAGTAC) were synthesized and HPLC/PAGE purified by Sigma. The sequence for the 80 mer Probe was selected from the pUC19 vector (108-187).
In vitro amplification
PCR reactions were performed on pUC 19C plasmid using primer pair Puc_UP125/Puc_Do168 for amplification of an 80-bp fragment. Amplification was performed in a total volume of 50 μL, containing: 5 ng pUC19C plasmid (New England Biolabs), 1.25 Unit Pfu DNA polymerase using Pfu buffer with MgSO4 (Fermenta), 0.1 mM dNTPs and 20 pmol of each primer. The PCR was carried in a GeneAmp PCR system (Perkin Elmer). Thermal Cycling for the PCR primer pair Puc_UP125/Puc_Do168 was performed with initial denaturation at 95°C for 60 s followed by 35 cycles (denaturation 95°C for 60s, annealing 58°C for 30 s and extension at 72°C 120 s) and a final extension 72°C for 5 min [19].
Preparation of ssDNA template
Streptavidin-coated super paramagnetic beads (300 μg) (Dynabeads M-280-streptavidin, Invitrogen) were washed three times by washing buffer supplied from the manufacturer. The biotinylated PCR product (50 L) was added to the washed beads and the solution was incubated for 30 min at 43°C. Unbound DNA was removed by washing three times with 100 μL of washing buffer. NaOH (5 μL 0.10 M) was added to the beads and the solution was incubated for 3 min. The supernantant (ssDNA) was removed and diluted in acetic acid (5 μL 0.10 M).
DNA ligation
The DNA concentration for the ligase reaction was 0.5 μM. The samples were incubated with 0.05 Units/μL to 5 Units/μL of CircLigase™(Epicenter) in CircLigase buffer, including 50 μM ATP and 2.5 mM MnCl2, at a total volume of 20 μL. The samples were incubated for different time periods.
DNA priming
The primers (10 pmol) were added to three micro liters of ligated probe diluted in a volume of 10 μL annealing buffer from Qiagen and hybridized at 80°C for 1 min.
Gel electrophoresis
Samples were analyzed by denaturing polyacrylamide gel electrophoresis (15% TBE-Urea Gels) (Bio-Rad). For visualization of DNA, gels were stained with Gelred (Biotium).
Enzymtic preparation of the ligated product
Ten μl of the product from the ligation reaction (se above) was incubated with exonuclease I (10 U, New England Biolabs), alkaline phosphatase (1U, Roche) and single stranded DNA binding protein (2 μg) at 37°C for 60 min. Inactivation of enzymes was carried out at 98°C for 3 min.
Ligase assay
The ligation efficiency was determined by DNA sequencing (Pyrosequencing) over the ligation site. The sequencing was performed at 30°C in a volume of 50 μl on an automated PyroMark Q96 system (Qiagen). Briefly, 10 μl of a primed ligation reaction (as described above) were used for an assay. Primed target DNA was added to the 30 μl Pyromark annealing buffer (Qiagen). The reaction was started by automatic addition of enzymes (Qiagen) and substrates (Qiagen). The sequencing procedure was carried out by stepwise elongation of the primer-strand upon sequential addition of the different deoxynucleoside triphosphates (Qiagen) and simultaneous degradation of nucleotides by apyrase.
Results
Principle of the ligase assay
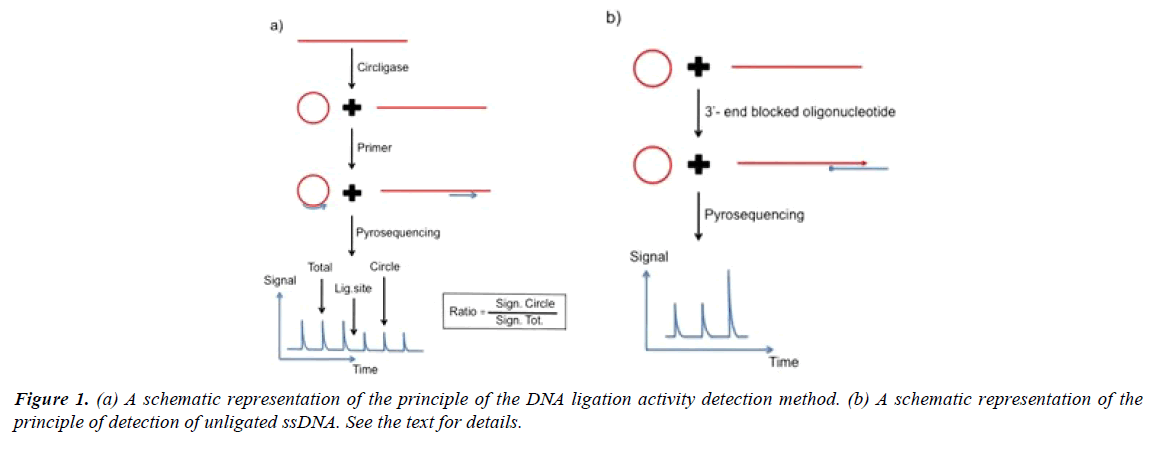
A schematic representation of the principle of the method for detection of ligase activity is shown in Figure 1. Singlestranded DNA is incubated with DNA ligase for a specific time period. After addition of detection primer and heating, the sample is cooled. In the schematic example the primer anneals at a position three bases before the ligation site. The DNA sequence directly after the primer is determined by directed nucleotide addition. The height of the signal from the base incorporation prior the ligation site represents the total amount of primed DNA (ligated plus unligated DNA). The height of the signal after the ligation site represents ligated ssDNA. By comparing the signal strength from the nucleotide incorporation event after the ligation site with the strength of the signal prior the ligation site an estimate of the ligation efficiency is obtained. In the example three bases before and three bases after the ligation site are analyzed.
The robustness of the assay can be further improved by exact estimation of the amount of unligated products. As illustrated in Figure 1a and 1b 3’-end blocked oligonucleotide that hybridize with both the unligated DNA and the ligated circulized DNA can function as template for DNA polymerase catalyzed nucleotide incorporation. The unligated ssDNA function as primer for the DNA polymerase reaction. The signal height can be used as an estimate of the exact amount of unligated DNA. The added blocked oligonucleotide cannot function as a sequencing primer on either ligated or unligated DNA as it is blocked in the 3’- end. In the example four bases are analyzed. The last signal represents a homopolymer region of two identical bases. For exact estimation of the amount of ligated product, as well as, unligated product, an internal inorganic pyrophosphate standard can be added after the nucleotide incorporation reactions.
Detection of ligase activity
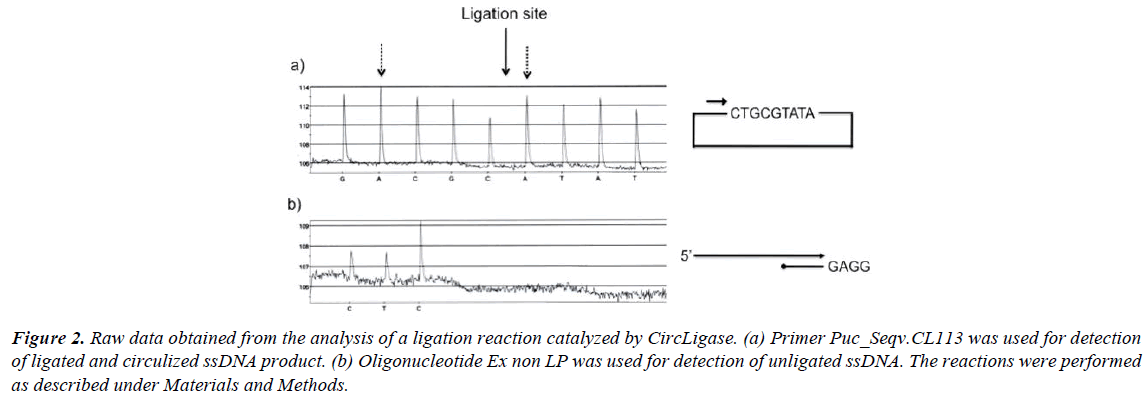
In Figure 2, raw data from experiments performed with CircLigase are shown. An 80-mer-long oligonucleotide (5’-end phosphorylated) was incubated with CircLigase for 1 hour. The ligation efficiency was determined by the method described above. The yield of ligation can be estimated by comparing the height of the first nucleotide incorporation (dATP) after the ligation site with the height of the second nucleotide incorporation (dATP) prior the ligation site (Figure 2a). From the comparison a yield of about 90% can be calculated. It is worth noting that incorporation of nucleotide A always gives a slight higher (10%) signal than the other three nucleotides (19, Qiagen). Of that reason it is practical to compare similar bases before and after the ligation site (C, G and T can always be compared with each other). The uneven signals observed in the described experiment, is due to frayed ends of the oligonucleotide used. This is normal for such a long oligonucleotide [20]. This phenomenon is further described below.
Figure 2: Raw data obtained from the analysis of a ligation reaction catalyzed by CircLigase. (a) Primer Puc_Seqv.CL113 was used for detection of ligated and circulized ssDNA product. (b) Oligonucleotide Ex non LP was used for detection of unligated ssDNA. The reactions were performed as described under Materials and Methods.
The ligation efficiency can also be analyzed with the second approach as described above (Figure 1b). In Figure 2b the amount of unligated product is estimated by the use of a specifically designed 3’-end blocked oligonucleotide. From the illustrated experiment it can be estimated that about 10% of the probe was not circulized.
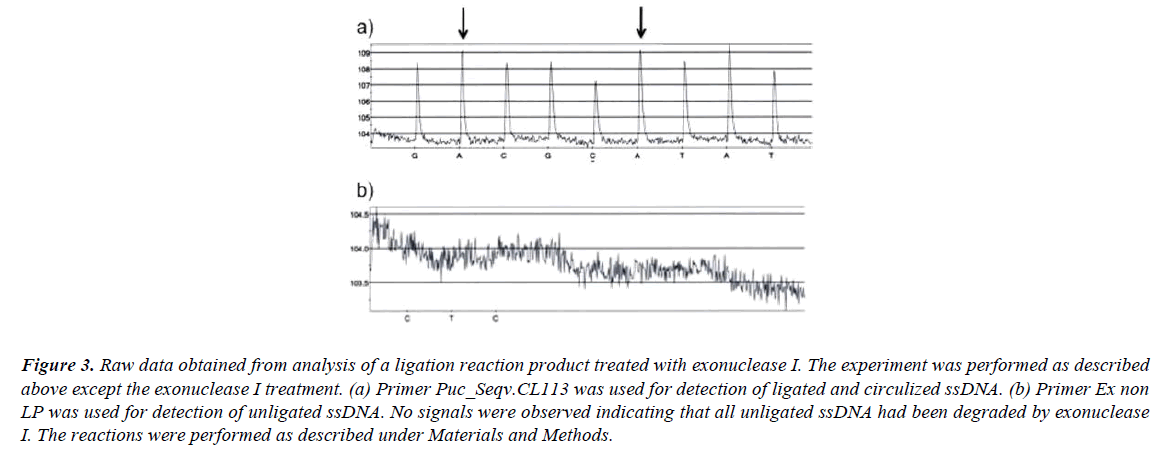
If the ligation reaction mixture (after the ligation reaction is finished) is treated with exonuclease I all unligated oligonucleotides are eliminated and no signals are observed (Figure 3b). Exonuclease I catalyze removal of nucleotides from ssDNA in the 3’ to 5’ direction. After the exonuclease I treatment the signal strengths from the first nucleotide incorporation (dATP) after the ligation site and the second nucleotide incorporation (dATP) prior the ligation site are equal (Figure 3a). This later result further strengthen the fact that all unligated ssDNA have been eliminated by the exonuclease I treatment.
Figure 3: Raw data obtained from analysis of a ligation reaction product treated with exonuclease I. The experiment was performed as described above except the exonuclease I treatment. (a) Primer Puc_Seqv.CL113 was used for detection of ligated and circulized ssDNA. (b) Primer Ex non LP was used for detection of unligated ssDNA. No signals were observed indicating that all unligated ssDNA had been degraded by exonuclease I. The reactions were performed as described under Materials and Methods.
Frayed ends
The uneven signals observed in the raw data shown in Figure 2a indicate that the ends of the used oligonucleotide are frayed, which probably is due to that the used oligonucleotide is a mixture of oligonucleotides of different lengths. It is well known that longer oligonucleotides are more difficult to purify than shorter [20].
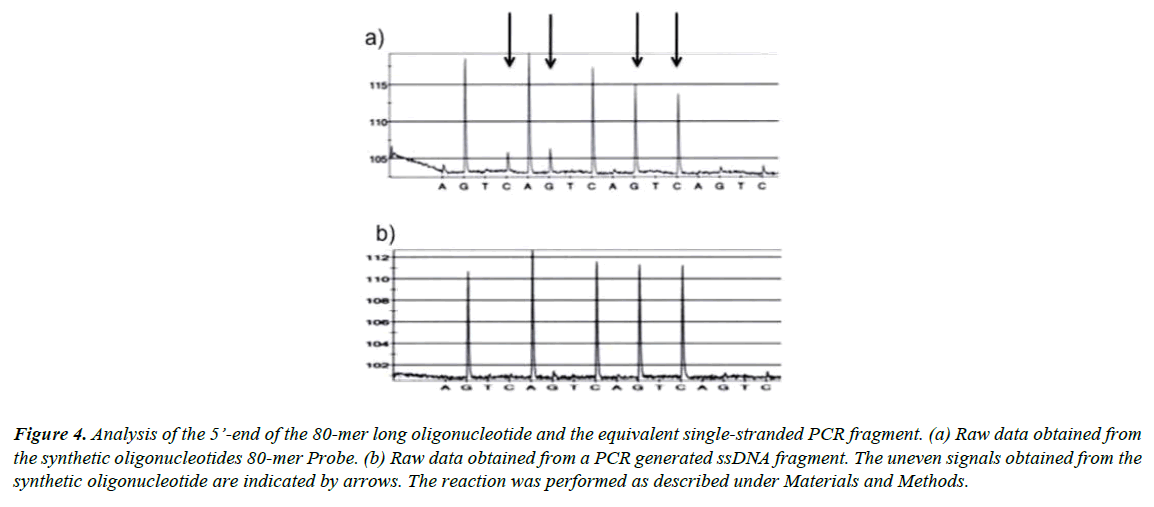
To further test the idea that the uneven signals were due to a heterogeneous population of oligonucleotides containing truncated oligonucleotides we prepared an equivalent PCR fragment. Thereafter, the five last bases at the 5’-end of both the synthetic 80-mer long oligonucleotide and the equivalent PCRamplified template were sequenced. From the raw data shown in Figure 4 it is clear that the quality of the PCR product (Figure 4b) was superior to the quality from the synthetic DNA (Figure 4a). The small signals shown in the trace (Figure 4a) indicate that the 80-mer-long synthetic oligonucleotide was impure and frayed in the 5’-end.
Figure 4: Analysis of the 5’-end of the 80-mer long oligonucleotide and the equivalent single-stranded PCR fragment. (a) Raw data obtained from the synthetic oligonucleotides 80-mer Probe. (b) Raw data obtained from a PCR generated ssDNA fragment. The uneven signals obtained from the synthetic oligonucleotide are indicated by arrows. The reaction was performed as described under Materials and Methods.
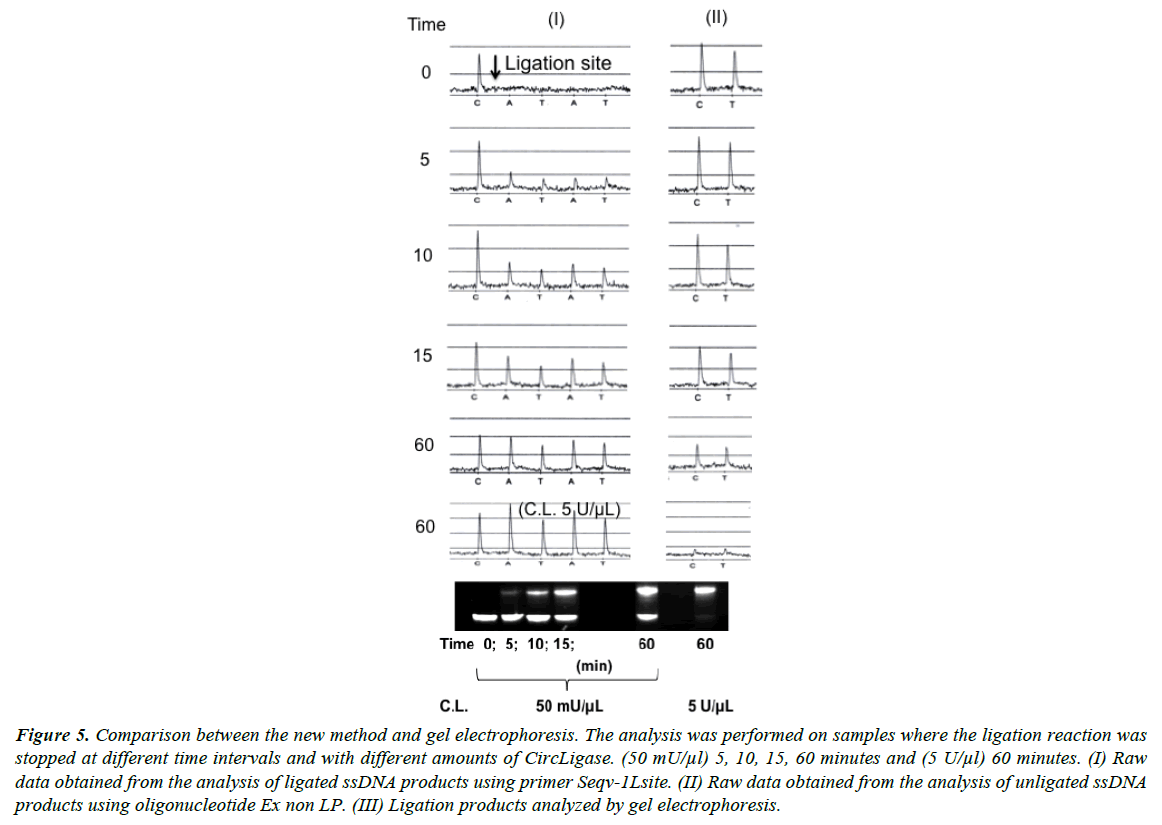
Comparison with gel electrophoresis
The standard procedure for detection of a ligation reaction is gel electrophoresis. In Figure 5 results obtained by the new method are compared with results from traditional electrophoresis. Both methods show similar results from experiments performed with different amounts of DNA ligase and with different incubation times. Close to 100% ligation efficiency was obtained with 100 units CircLigase incubated with 10 pmol oligonucleotide for 60 minutes.
Figure 5: Comparison between the new method and gel electrophoresis. The analysis was performed on samples where the ligation reaction was stopped at different time intervals and with different amounts of CircLigase. (50 mU/μl) 5, 10, 15, 60 minutes and (5 U/μl) 60 minutes. (I) Raw data obtained from the analysis of ligated ssDNA products using primer Seqv-1Lsite. (II) Raw data obtained from the analysis of unligated ssDNA products using oligonucleotide Ex non LP. (III) Ligation products analyzed by gel electrophoresis.
Discussion
Circulized ssDNA is used in many different applications. In linear rolling circle amplification (LRCA), circular probes serves as templates for DNA polymerases generating numerous concatemirized copies of the circle [21,22]. Exponential signal amplification can be obtained if the circular probe is combined with a pair of sense and antisense primers [23,24]. Circulized ssDNA is central for signal amplification of padlock probes [25,26] and for sensitive protein analyses [27,28].
For most applications it is important to optimize the ligation step (the circulization of ssDNA) to obtain as high as possible efficiency. The quality of the circles is also of importance for many applications where specific interactions are studied. With the new method we have studied circulization of both short and long oligonucleotides (30 mers to 80 mers). The circles were circulized utilizing CircLigase I and II. We were also able to detect frayed ends of the long oligonucleotides. By utilizing PCR for production of ssDNA much better quality was obtained.
The new method has been used for analysis of ssDNA circulization but can in principle be used for analysis of dsDNA ligation. To be able to detect dsDNA ligation a blocked oligonucleotide complementary to the splint oligonucleotide can be used in a similar way as earlier described for preparation of dsDNA for sequencing [29].
In conclusion, we have described an alternative procedure for detection of the ligation reaction and for analysis of the quality of the ligated product. The described procedure is simple and fast with a low number of steps and short hands-on times. The simplicity of the procedure makes it a particularly suited alternative to the traditionally used gel electrophoresis method. The new method is unique because of the possibility to analyze DNA ligation activity and DNA quality simultaneous. The DNA quality is of most importance for applications where specific DNA sequences, mutations and polymorphisms are analyzed. The new method has the potential to be a useful tool for researchers working with both basic and applied projects in the field of biotechnology.
Acknowledgements
This work was supported by grants from the European Commission Seventh framework programme (HiPerDART).
References
References
- Tomkinson AE, Vijayakumar S, Pascal TJM. Ellenberger, DNA ligases: structure, reaction mechanism, and function. Chem Rev 2006;106:687-99.
- Shuman S, Lima CD. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. CurrOpinStructBiol2004;14:757-64.
- Wilkinson A, Day J, Bowater R. Bacterial DNA ligases. Mol Microbial2001;40:1241-48.
- Lehman IR. DNA ligase: structure, mechanism, and function. Science 1974;186:790-97.
- Cao W. DNA ligases and ligase-based technologies. ClinApplImmunol Rev 2001;2: 33-43.
- Fredriksson S,Gullberg M,Jarvius J, et al. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol 2002;20:473-77.
- Cao W. Recent developments in ligase-mediated amplification and detection. Trends Biotechnol2004;22:38-44.
- Landegren U, Kaiser R, Sanders J,et al. A ligase-mediated gene detection technique. Science 1988;241:1077-80.
- Barany F. Genetic disease detection and amplification using cloned thermostable ligase. ProcNatlAcadSci USA 1991;88:189-93.
- Baron H, Fung S,Aydin A, et al. Oligonucleotide ligation assay (OLA) for the diagnosis of familial hypercholesterolemia. Nature Biotechnol 1996;14:1279-82.
- Dunn JJ, Butler-Loffredo LL, Studier FW. Ligation of hexamers on hexamer templates to produce primers for cycle sequencing or the polymerase chain reaction. Anal Biochem 1995;228:91-100.
- Kaczorowski T,Szbalski W. Assembly of 18-nucleotide primers by ligation of three hexamers: sequencing of large genomes by primer walking. Anal Biochem 1994;221:127-35.
- Olivera BM, Lehman IR. Linkage of polynucleotides through phosphodiester bonds by an enzyme from Escherichia coli. Proc Nat AcadSci USA 1967;57:1426-33.
- Modrich P, Lehman IR. Enzymatic joining of polynucleotides IX. A simple and rapid assay of polynucleotide joining (ligase) activity by measurement of circle formation from linear deoxyadenylate-deoxythymidylate copolymer. J BiolChem 1970;245:3626-31.
- Goodchild J,Vishwanatha JK. A novel assay for DNA ligase. Nucleic Acids Res 1991;19:3745.
- Courtney BC, Williams KC, Bing QA, et al. Capillary gel electrophoresis as a method to determine lgation efficiency. Anal Biochem1995;228:281-86.
- Liu L, Tang ZW, Wang KM, et al. Using molecular beacon to monitor activity of E.coli DNA ligase, Analyst 2005;130:350-57.
- Vacek J, Cahova K, Palecek E, et al. Label-free electrochemical monitoring of DNA ligase activity. Anal Chem2008;80:7609-13.
- Eriksson J,Gharizadeh B, Nourizad N, et al. 7-Deaza-2'-deoxyadenosine-5'-triphosphate as an alternative nucleotide for the pyrosequencing technology, Nucleosides Nucleotides Nucleic Acids 2004;23:1583-94.
- Temsamani J, Kubert M,Agrawal S. Sequence identity of the n-1 product of a synthetic oligonucleotide. Nucleic Acids Research 1995;23:1841-44.
- Fire A,Xu SQ. Rolling replication of short DNA circles.ProcNatlAcadSci 1995;92:4641-45.
- Liu D,Daubendiek SL, Zillman MA, et al. Rolling Circle DNA Synthesis: Small Circular Oligonucleotides as Efficient Templates for DNA Polymerases. J Am ChemSoc1996;118:1587-94.
- Lizardi P, Huang X, Zhu Z, et al. Nat Genet 1998;19:225-32.
- Zhang DY,Brandwein MY,Hsuih TC, et al. Amplification of target-specific, ligation-dependent circular probe. Gene 1998;211:277-85.
- Nilsson M, Malmgren H,Samiotaki M, et al. Padlock probes: circularizing oligonucleotides for localized DNA detection. Science 1994;265:2085-88.
- Baner J, Nilsson M, Mendel-Hartvig M, et al. Signal amplification of padlock probes by rolling circle replication. Nucl Acids Res 1998;26:5073-78.
- Schweitzer B, Wiltshire S, Lambert J, et al. Ward, Immunoassays with rolling circle DNA amplification: a versatile platform for ultrasensitive antigen detection.ProcNatlAcadSci 2000;97:10113-119.
- Soderberg O, Leuchowius KJ, Gullberg M, et al. Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods 2008;5:227-32.
- Nordstrom T, Alderborn A,Nyren P. Method for one-step preparation of double-stranded DNA template applicable for use with Pyrosequencing technology. J BiochemBiophys Methods 2002;52:71-82.