- Biomedical Research (2012) Volume 23, Issue 2
Large-Scale Production of Highly Active Recombinant Omi from Escherichia coli Expressed Soluble Protein Aggregates
Md. Mashiar Rahman1#*, Shahina Akhter1#, Dipendra Raj Pandeya1,2, Roshan D’Souza1 and Seong-Tshool Hong1
1Department of Microbiology and Genetics and Institute for Medical Science, Chonbuk National University Medical School, Jeonju 561-712, South Korea.
2Department of Biochemistry, Nepalese Army Institute of Health Sciences, Sanobharayang, Kathmandu, Nepal.
#These authors contributed equally to this work.
- *Corresponding Author:
- Md. Mashiar Rahman
Department of Microbiology and Genetics and Institute for Medical Science
Chonbuk National University Medical School
Jeonju 561-712, South Korea
Accepted Date: February 24 2012
Abstract
Recent increasing evidences have linked Omi to neuroprotection and the cellular protein quality control system. However, it is very difficult to produce functionally active Omi to study the role of Omi in vitro. Here, we report a method for mass production of functionally active high quality Omi through heterologous Escherichia coli expression. The heterologously expressed Omi was present mostly in soluble cell fraction rather than insoluble cell fraction as inclusion bodies, but the protein in soluble cell fraction had very poor activity. In this method, low concentration of denaturing agents was added to the lysate for complete solubilization and monodispersion of Omi after lysising Escherichia coli. The solubilized and monodisperesed protein sample was purified by Ni-NTA agarose affinity chromatography at denatured condition. The purified denatured Omi was then refolded by a rapid-dilution in an optimized protein refolding buffer that was designed by systematic variation of parameters favoring the refolding, preventing aggregate formation and influencing the stability of the folding intermediates. Around 20 mg of properly folded Omi was obtained from 1 liter of bacterial culture by this method which is the highest production reported so far. The biological activity was confirmed by its ability to degrade β-casein. This method is a straightforward and efficient purification procedure to obtain a higher yield of proper biologically active Omi and that Omi can be used for the investigation of Omi’s association with neuroprotection in various neurodegenerative diseases.
Keywords
Omi, soluble protein aggregates, denature, solubilization, monodispersion, purification, refolding, activity
Introduction
The HtrA family of serine proteases, whose members have been identified in most eukaryotes and prokaryotes, takes part in cells’ protection against stress conditions including heat shock, oxidative stress, inflammation, ischemia/reperfusion, and cancer [1]. The most extensively studied member of this family is the E. coli HtrA/DegP that acts as a serine protease degrading misfolded proteins in the periplasm at high temperatures and as a molecular chaperone at low temperatures [2]. To date, four human homologues, named HtrA1-4 of HtrA have been identified. The common feature of the HtrA family of proteins is a highly conserved trypsin-like serine protease domain and one or two PDZ domains [1]. HtrA2 also known as Omi is the only member of the family of four mammalian HtrA proteases that has a mitochondrial localization signal in N-terminus and auto catalytically processed into mature form to reveal a Reaperlike amino-terminal motif [3]. In vitro studies showed that Omi is an apoptosis-inducing mitochondrial protein [4, 5]. The neuroprotective role of Omi has been initially identified by the demonstration of neurodegeneration in mice lacking HtrA2 expression or function [6,7]. Like Alzheimer's disease-associated proteins, presenilin-1 and oligomeric amyloid-β [8,9], fibrilized α-synuclein and oligomeric prion are thought to be associated with Omi. Therefore, the recombinant production of high quality Omi in substantial quantity is important for its biological and structural studies.
E. coli are regarded as an outstanding factory for recombinant expression of proteins. Heterologously expressed recombinant proteins in E. coli result in either soluble cell fraction or insoluble inclusion bodies [10,11]. Although may not frequent, some proteins do occur both in soluble and insoluble cell fractions [10, 12], and some proteins do occur as soluble aggregates which are ranging from particulate material to fibril-like structures [13,14]. The general belief that soluble protein species are properly folded and fully functional in contrast to the insoluble inclusion bodies [11,15]. However, current research findings that the solubility is not a sufficient criterion to ensure that proteins are properly folded because the solubility and conformational quality in recombinant bacteria are divergently controlled [16,17]. Reducing the growth temperature of recombinant E. coli cells from 37°C to 16°C results in a significant increase of the specific emission of a recombinant green fluorescent protein (GFP) [18], indicating that at 37°C an important fraction of soluble species have not maturated to an optimal conformation for fluorescence emission. The finding of the so-called soluble aggregates, which are fibril-like structures, in E. coli cells overproducing GFP variants, indicates that even soluble species can display an amyloidal organization, and that GFP in these soluble aggregates may eventually fold into a form very different from the native conformation [14,19]. However, it was suggested that a better quality criterion of protein would be that the preparation of the protein of interest should be structurally homogeneous and mono disperses [14,16].
In this work, we describe an efficient and simple method to purify a large amount of quality Omi by using intermediate concentration of denaturing agents followed by refolding in an optimized protein refolding buffer. The refolded Omi shows a great promising proteolytic activity by degrading a generic substrate β-casein.
Materials and Methods
Protein, protease inhibitor, antibodies and buffers
Beta-Casein (Cat# C6905) was purchased from Sigma- Aldrich, St. Louis, MO. The primary rabbit polyclonal anti-Omi antibody (Cat# AF1458) was purchased from R & D System and the secondary horseradish peroxidaseconjugated goat anti-rabbit IgG (H + L) antibody (Cat# W4021) was purchased from Promega Corporation. Protease inhibitor cocktail, Complete mini from Roche Applied Bioscience. The protein expression, purification, refolding and dialysis were performed by using different buffers: native cell lysis buffer A (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2.5 mM EDTA, 0.5% Triton X-100, 4 mM MgCl2, 0.5 mg/mL lysozyme and protease inhibitor cocktail Complete mini), native washing buffer B (50 mM Tris-HCl, pH 8.0; 150 mM NaCl, 1.0% Triton X-100, 5.0 mM EDTA, and protease inhibitor Complete mini), protein denature and purification buffer C (50 mM Tris-HCl, pH 8.0; 5 M Urea, and 300 mM NaCl), protein solubilization and monodispersion buffer D (50 mM Tris-HCl, pH 8.0; 10 M Urea, 500 mM NaCl and 20 mM Imidazole, pH 6.0), optimized protein refolding buffer E (50 mM Tris- HCl, pH 8.5; 500 mM NaCl, 5 mM EDTA, 1 mM GSH, 0.1 mM GSSG, 500 mM Arginine and 15% Glycerol or sucrose) and dialysis/Omi buffer F (20 mM HEPES, pH 7.5 and 100 mM NaCl).
Construction of expression vector
Corresponding DNA sequence to human recombinant mature Omi (134-458) was cloned in-frame in the bacterial expression vector pET28a+ (Novagen, 69864-3) under six His residues at the N-terminal to simplify the purification procedures as previously described [20]. Briefly, the cDNA encoding mature Omi was amplified by PCR using the following primers: sense primer A, 5′-GTC CTC GCC CAT ATG GCC GTC CCT AGC C-3′; antisense primer B, 5′-G GCT CTC GAG TCA TTC TGT GAC CTC AGG G-3′ from a full-length cDNA clone of human Omi in the pCMV-SPORT6 vector (Image 97002RG) which was obtained from Invitrogen (USA). The PCR product was digested with NdeI/XhoI (New England Biolabs Inc.) and subcloned into the NdeI/XhoI sites of the pET28a+ expression vector. The construct obtained was confirmed by colony PCR, restriction analysis and DNA sequencing. It was propagated in strain DH5α (RBC, RH617 H) and purified with a Nucleogen Plasmid Mini Kit (Nucleogen, P3114), then transformed into E. coli BL21 (DE3) pLysS (Stratagene, 200132- 41) by Bio-Rad Gene Pulser II apparatus by using 0.2 cm cuvette (Bio- Rad, 1652086) for bacterial expression of Omi.
Omi expression
In order to express His-tag Omi, a single colony of successfully transformed E. coli BL21 (DE3) pLysS was grown overnight at 37°C with continuous shaking in 25 mL Luria Bertani (LB) medium supplemented with 50 μg/mL kanamycin at 250 rpm as a starter culture. 1.0 mL of overnight culture was then inoculated to fresh 100 mL LB medium containing 50 μg/mL kanamycin and growth was continued at 37°C under continuous shaking at above mentioned rpm. To optimize the culture conditions, Omi expression was monitored by culturing the bacteria until the absorbance at 600 nm reached at 0.6, 0.8 and 1.0 prior and post induction with 0.5 and 1.0 mM Isopropyl β-D-1- thiogalactopyranoside (IPTG) (Duchefa Biochemie, I1401.0005) for 2 to 6 h followed by withdrawn of 1.0 mL of culture samples at A600 of 0.6, 0.8, and 1.0 prior and post induction with 0.5 and 1.0 mM IPTG. The degree of Omi expression was evaluated by SDS-PAGE (Data not shown) and western blot analysis (Fig. 1). After optimization of small-scale expression, large-scale expression of Omi was carried out by culturing the bacteria at 37°C to mid-log phase of absorbance at 600 nm reached at 0.8 followed by induction with 0.5 mM IPTG for 5 h at 20°C. After induction, the bacterial cells were harvested by centrifugation at 6000× g for 15 min and stored at either -20ºC or at -80ºC till further use.
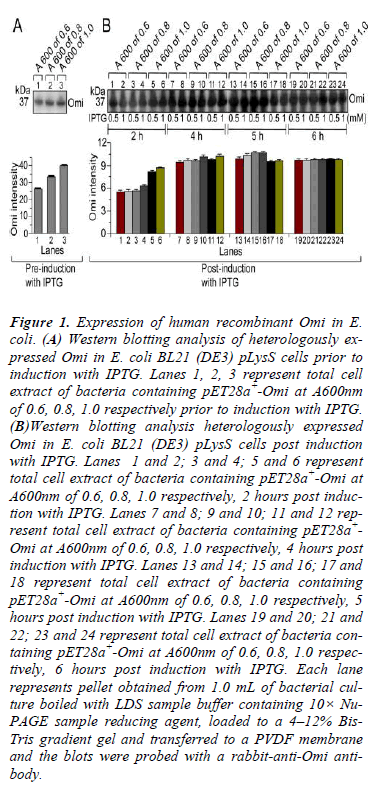
Figure 1: Expression of human recombinant Omi in E. coli. (A) Western blotting analysis of heterologously expressed Omi in E. coli BL21 (DE3) pLysS cells prior to induction with IPTG. Lanes 1, 2, 3 represent total cell extract of bacteria containing pET28a+-Omi at A600nm of 0.6, 0.8, 1.0 respectively prior to induction with IPTG. (B)Western blotting analysis heterologously expressed Omi in E. coli BL21 (DE3) pLysS cells post induction with IPTG. Lanes 1 and 2; 3 and 4; 5 and 6 represent total cell extract of bacteria containing pET28a+-Omi at A600nm of 0.6, 0.8, 1.0 respectively, 2 hours post induction with IPTG. Lanes 7 and 8; 9 and 10; 11 and 12 represent total cell extract of bacteria containing pET28a+- Omi at A600nm of 0.6, 0.8, 1.0 respectively, 4 hours post induction with IPTG. Lanes 13 and 14; 15 and 16; 17 and 18 represent total cell extract of bacteria containing pET28a+-Omi at A600nm of 0.6, 0.8, 1.0 respectively, 5 hours post induction with IPTG. Lanes 19 and 20; 21 and 22; 23 and 24 represent total cell extract of bacteria containing pET28a+-Omi at A600nm of 0.6, 0.8, 1.0 respectively, 6 hours post induction with IPTG. Each lane represents pellet obtained from 1.0 mL of bacterial culture boiled with LDS sample buffer containing 10× Nu- PAGE sample reducing agent, loaded to a 4–12% Bis- Tris gradient gel and transferred to a PVDF membrane and the blots were probed with a rabbit-anti-Omi antibody.
Western blot
The pellet obtained from 1.0 mL of bacterial culture resuspended in 1 mL PBS, and aliquots of the representative samples were mixed with NuPAGE 4× LDS sample buffer (Invitrogen, Carlsbad, CA, USA) and 10× Nu- PAGE sample reducing agent (Invitrogen, Carlsbad, CA, USA) in a total reaction volume of 30 μL. Samples were boiled for 10 min at 70ºC and 10 μL was loaded to a 4- 12% Bis-Tris gradient gel (Invitrogen, Carlsbad, CA, USA) and subjected to electrophoresis by MES running buffer system (Invitrogen, Carlsbad, CA, USA). After electrophoresis, the protein was transferred to a PVDF membrane (Bio-Rad Laboratories, Harcules, CA) and blocked with 5% non-fat dry milk (Bio-Rad Laboratories, Harcules, CA) in PBST-Triton X-100 (PBS with 0.05% Tween-20 and 0.1% Triton X-100) for 3 h at room temperature. After 5 min washing with PBST- TritonX-100, the membrane was incubated overnight at 4ºC with a rabbit anti-Omi antibody in an antibody dilution buffer at a concentration of 0.25μg/mL with gentle rocking. The membrane was washed 4 times, each of with 10 min by PBST TritonX-100 with gentle rocking. Following washing, the membrane was incubated for 1 h at room temperature with a horseradish peroxidase-conjugated goat anti-rabbit IgG (H + L) antibody. Thereafter, the membrane was washed similarly as primary antibody washing and the chemiluminescent substrate, Immun-StarTM Western CTM Kit (Bio-Rad Laboratories, Harcules, CA), was applied to the PVDF membrane. The images were captured with a Chemi-doc XRS camera equipped with a Bio-Rad Quantity One imaging system. The stock solutions of primary and secondary antibodies were diluted in an antibody dilution buffer (0.5% BSA in PBST-Triton X-100).
Nature of Omi expression
Since we have experienced of very poor activity of native Omi, we washed the harvested bacterial pellet once with PBS and resuspended in native cell lysis buffer A followed by incubation at 4ºC for 30 min. Thereafter, the cell suspension was subjected to sonication for 15 × 10 sec, 6 bursts at high intensity with 1 min cooling period on ice (Bandelin Sonopuls HD 2070, Berlin, Germany). Following sonication, the bacterial cell lysate was centrifuged at 20,000× g for 45 min at 4ºC. The supernatant was saved and the pellet was resuspended in native washing buffer B and centrifuged again at the same condition to wash the pellet. The supernatant was saved and the washed pellet was resuspended in denaturing buffer C and allowed to solubilize for monodispersion overnight at room temperature or 4ºC with constant stirring on a magnetic stirrer. The overnight solubilized sample was centrifuged again at the same condition mentioned above to remove if any precipitation. Aliquots of supernatant after centrifugation of cell lysate in buffer A, wash of pellet in buffer B and solubilized pellet in buffer C were electrophoresed and stained with Commassie blue to check the nature of expression of Omi in E. coli.
Purification of Omi at denatured condition
The harvested bacterial pellet obtained from 1 L culture was washed once in PBS and pelleted by centrifugation at 6000× g for 15 min. This pellet was resuspended in 20 mL of native cell lysis buffer A and incubated at room temperature for 30 min and subjected to sonication for 15 × 10 sec, 6 bursts at high intensity with 1 min cooling period on ice (Bandelin Sonopuls HD 2070, Berlin, Germany). Following sonication, buffer D was added to 20 mL bacterial cell lysate to make a final concentration half of original concentration of each constituent in buffer D and was sonicated again for 10× 10 sec, 6 bursts at high intensity with 1 min cooling period on ice. Thereafter, the bacterial cell lysate was centrifuged at 20,000× g for 45 min at 4ºC and the supernatant was allowed to solubilize for monodispersion overnight at room temperature or 4ºC with constant stirring on a magnetic stirrer. The solubilized protein solution was centrifuged at 20,000× g for 45 min to remove if any unsolubilized material. Pipetted of 5.0 mL of Ni-NTA agarose (Invitrogen, R901-01) into an econo-pac chromatography column (Bio-Rad, 732-1010) and it was prepared for protein purification according to manufacturer instruction. Twenty mL of supernatant was allowed to bind for 1 h at 4ºC with gentle agitation to a 5- mL Ni-NTA agarose column pre-equilibrated with 30 mL of buffer C containing 10 mM Imidazole (pH 6.0). After binding, the supernatant was allowed to pass through and the resin was washed twice, each of with 15 mL of buffer C containing 10 mM Imidazole (pH 6.0) at 4ºC for 10 min with gentle agitation. Thereafter, the resin was washed 5 times, each of with 15 mL of buffer C containing 50 mM Imidazole (pH 6.0) at 4ºC for 10 min with gentle agitation to remove the impurities devoid of the histidine tag. The histidine-tag denatured Omi was then eluted as about 1.5 mL fractions with three column volumes of buffer C containing 500 mM Imidazole (pH 6.0). Aliquots of each Ni- NTA fractions were subjected to SDS-PAGE analysis to check the purity and comparative bulk protein fractions.
Size exclusion chromatography
All eluted Ni-NTA fractions with OD280 reading equal or above 0.1 were combined. The protein was re-purified by the PD-10 Sephadex® G-25M Column (Sigma, 54805) pr-equilibrated with buffer C to remove lower molecular weight fragments. The protein was eluted with the same buffer followed by measuring the protein concentration by the Bradford assay kit (Pierce, Rockford, USA). The purity of the re-purified Omi was checked by SDS-PAGE and Comassie blue staining.
Refolding of denatured Omi
The re-purified Omi was reduced by adding dithiothreitol (DTT) to a final concentration of 10 mM at 37ºC for 1 h with continuous shaking at 25 rpm on an Intelli-Mixer (Seoulin Bioscience Co., Ltd., Seoul, South Korea). This reduced solution was filtered through a 0.45 μm filter (Sartorious Stedim Biotech, 6555) to remove if any precipitation. Refolding of recombinant Omi was then assessed using a series of buffers differing in pH, concentration of redox couple (reduced glutathione, GSH to oxidized glutathione, GSSG), aggregation suppressors (LArginine, NaCl, Triton X-100 and Urea) and co-solvents (glycerol and sucrose) by using a rapid-dilution method so as to get maximum amounts of refolded protein. Thereafter, denatured protein was refolded in an optimized protein refolding buffer E by maintaining the final protein concentration at 40-50 μg/mL with constant stirring at 4ºC for 24 h. Aliquot of diluted refolded Omi was checked by SDS-PAGE and Comassie blue staining. Refolded Omi was centrifuged at 20,000× g for 1 h at 4ºC to remove if any precipitation. Soluble, refolded protein was then concentrated at 4ºC on an Amicon stirred cell ultrafiltration unit using an YM-10 membrane (Millipore Corporation, Billerica, USA). Aliquot of concentrated refolded Omi was analyzed by SDS-PAGE and Comassie blue staining to check the purity and concentration. Further purification and dialysis of refolded Omi was performed on a PD-10 column equilibrated with dialysis/Omi buffer F. The protein was eluted using the same buffer, and the protein concentration was measured and the protein was stored at -80ºC in 20% glycerol. Aliquot of dialyzed refolded Omi was also analyzed by SDS-PAGE and Commassie blue staining to check the purity and concentration.
Omi-proteolysis activity assay
The proteolytic activity of refolded Omi was assayed by incubation of 4 μM β-casein with 2 μM Omi or Omi buffer F (as a control) supplemented with 50 mM sodium phosphate (NaP) buffer (pH 7.4) at indicated temperatures for 2 h. Samples were analyzed by SDS-PAGE and Coomassie blue staining.
SDS-PAGE analysis
Aliquot of 20 μL of the representative samples were mixed with NuPAGE 4× LDS sample buffer and Nu- PAGE 10× sample reducing agent in a total reaction volume of 30 μL. Samples were boiled for 5 min at 70ºC, and 10 μL was loaded to a 4-12% Bis-Tris gradient gel and subjected to electrophoresis by MES running buffer system. After electrophoresis, the protein was stained by Coomassie Brilliant Blue R-250 (Sigma, B0149).
Results
Protein expression
In order to determine the optimal conditions for higher level of Omi expression, the transformed E. coli BL21 (DE3) pLysS-pET28a+-Omi was cultured at 20ºC without IPTG (Fig. 1A) and with IPTG induction (Fig. 1B), and culture samples withdrawn at A600 of 0.6, 0.8, and 1.0 prior, and 2-6 h post induction with 0.5 mM and 1.0 mM IPTG. Western blot analysis of both types of culture samples indicated that the optimum time and IPTG concentration for higher level of Omi expression was 5 h along with 0.5 or 1.0 mM IPTG. We then performed large scale culture of E. coli BL21 (DE3) pLysS-pET28a+-Omi at 20ºC for 5 h by induction with 0.5 M IPTG. As shown in Fig. 5A, native purified Omi showed very poor proteolytic activity against β-Casein.
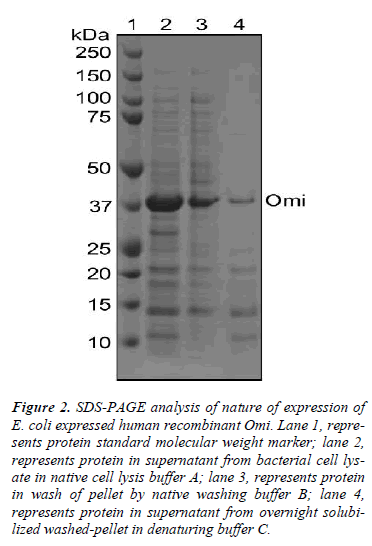
To understand this poorer functionality of Omi, we used different buffers to identify whether Omi is expressed as soluble or insoluble cell fractions as inclusion bodies. As shown in Fig. 2, the most Omi was observed in supernatant after high speed centrifugation of bacterial cell lysate in native cell lysis buffer A (lane 2). Washing of pellet by high concentration of detergent in native washing buffer B detached a large portion of Omi from remaining pellet from high speed centrifugation (lane 3). A very thin band was observed from overnight solubilization of the washed pellet by protein denaturing buffer C (lane 4). These results indicate that the most Omi is expressed as soluble cell fraction and that the soluble protein showed very poor proteolytic activity (Fig. 5A). Moreover, it was reported that the straightforward measuring the solubility of the protein is not a useful indicator of functional quality [14]. Thus, on the basis of the previous reports and our data, we conclude that Omi is expressed as soluble aggregates.
Figure 2: SDS-PAGE analysis of nature of expression of E. coli expressed human recombinant Omi. Lane 1, represents protein standard molecular weight marker; lane 2, represents protein in supernatant from bacterial cell lysate in native cell lysis buffer A; lane 3, represents protein in wash of pellet by native washing buffer B; lane 4, represents protein in supernatant from overnight solubilized washed-pellet in denaturing buffer C.
Protein purification and refolding
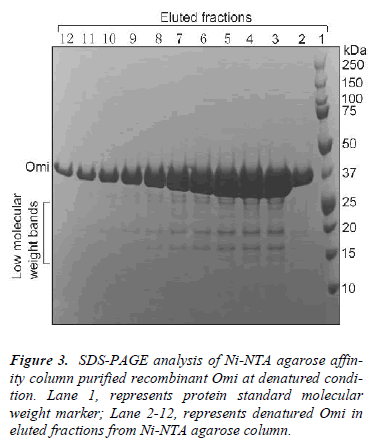
Recent increasing body of evidences clearly argues against a model picturing the soluble protein fraction as fully functional and structurally homogeneous because the specific biological activity rather than the presence of the protein species in the soluble cell fraction demonstrates the conformational quality of the product and, therefore, its biotechnological potential [14,16]. We used low concentration of denaturing agents to the lysed bacterial cells to solubilize and monodisperse the soluble protein aggregates for structural homogenization. After high speed centrifugation, the protein sample was purified under denaturing conditions by Ni–NTA agarose affinity chromatography as a single major band of about 37 kDa together with a few low molecular weight bands as determined by SDS-PAGE and Coomassie blue staining (Fig. 3).
To remove the low molecular weight bands, the protein eluted from Ni-NTA column was purified by size exclusion chromatography on a PD-10 Sephadex® G-25M column which provided a comparative highly purified protein as assessed by SDS-PAGE (Fig. 4, lane 2). After size exclusion chromatography, denatured Omi was refolded by a rapid-dilution method into an optimized protein refolding buffer E, yielding folded Omi (Fig. 4, lane 3). The refolding protocol for Omi was systematically standardized for optimum pH, temperature, ionic strength of buffer, requirement for various additives like redox pair, arginine, glycerol, and sucrose. It was noted that the denatured protein folded best at pH ~8.5 and at 4°C, while pH values above 8.9 and below 7.4 were unsuitable for refolding. It was also observed that the refolding process required inclusion of 0.5 M arginine, 5 mM GSH: 0.5 mM GSSG and 15% glycerol or sucrose. The feeding protein concentration, 40-60 μg/mL was crucial to achieve the best refolding of the protein. A concentration above 60 μg/mL resulted in a large amount of insoluble protein and low yield of the refolded protein. The diluted refolded omi was concentrated by Amicon stirred cell ultra filtration unit that helped also in removing low molecular weight fragments (Fig. 4, lane 4). A further purification of the refolded protein by size exclusion chromatography helped in removing impurities like improperly folded protein from the properly folded Omi (Fig. 4, lane 5). Thus 40 mg of protein was purified by Ni-NTA chromatography from 1 L of bacterial culture, which in turn provided 20 mg of refolded Omi.
Figure 4: SDS-PAGE analysis of re-purified denatured Omi and refolded Omi. Lane 1, represents protein standard molecular weight marker; Lane 2, PD-10 columnre- purified denatured Omi from Ni-NTA fractions; lane 3, diluted refolded Omi; lane 4, Amicon stirred cell ultrafiltration unit-concentrated refolded Omi; lane 5, PD-10 column-re-purified refolded Omi.
Proteolytic activity of Omi
In order to determine whether refolded Omi is properly folded, β-Casein was used to examine the proteolytic activity of recombinant Omi. The proteolysis activity of refolded Omi and native purified Omi was compared by temperature dependent degradation of β-Casein. Refolded Omi showed a great promising proteolytic activity both at normal and higher temperatures by complete degradation of β-Casein (Fig. 5B) whereas native purified Omi partially degraded β-Casein (Fig. 5A). These results indicated that Omi that we produced by denaturation and re naturation procedures was correctly folded because β- casein is a generic substrate for in vitro assay of the biological activity of mammalian heat-shock Omi [21].
Figure 5: Activity assay of purified recombinant Omi. (A) Activity assay of native Omi against β-Casein. (B) Activity assay of refolded Omi against β-Casein. Beta-casein was incubated with native Omi or refolded Omi or their respective buffer F supplemented with NaP buffer at the indicated temperatures for 2 h. The reaction mixtures were analyzed by SDS-PAGE analysis followed by Coomassie blue staining. NaP, sodium phosphate.
Discussion
The quality of recombinant proteins produced in bacteria and other host cells represents a major matter of concern in biological protein production and determines the potential use of target proteins for functional applications or structural analysis (11, 14). Because recombinant proteins in producing cells can be seen as adopting "a continuum of forms" expanding from soluble to insoluble cell fractions, and inclusion bodies as insoluble "clusters" of protein species (14, 15). In the present study, we reveal that over expression of Omi in E. coli cells results in soluble protein aggregates and that aggregation has been resulted from the acquaintance of proximate native structures that the proximate native structures shows poorer functionality. Thus, we develop a method of denaturation/ solubilization for monodispersion of soluble aggregates of Omi and its subsequent renaturation to repair the original functionality of Omi to regain of Omi’s proper biological activity with higher yield.
Although in general, inclusion bodies are solubilized by high concentration of denaturants in the presence of reducing agents; we used intermediate concentration of Urea without any reducing agents in lysis buffer to solubilize and monodisperse by denaturing the soluble protein aggregates. Because, Omi is not a disulfide bond containing protein [22], and 5 M Urea might be enough to achieve extensive binding sufficient to solubilize and monodisperse by denaturing the protein. Protein solubilization from the protein aggregates using high concentration of chaotropic agents result in the loss of secondary structure leading to the random coil formation of the protein structure and exposure of the hydrophobic surface. Loss of secondary structure during solubilization and the interaction among the denatured protein molecules during refolding resulting in their aggregation are considered to be the main reasons for the poor recovery of bioactive proteins from the protein aggregates. Moreover, the presence of a reducing agent in any step of protein purification may decrease the yield of the protein due to its autocleavage [23, 24].
In vivo folding of proteins is rapid and is assisted by folding catalysts, such as disulfide isomerase, and molecular chaperones. In vitro refolding of proteins into bioactive form is cumbersome due to improper disulfide bond formation, aggregation and instability of folding intermediates. Hence, it is crucial to identify the correct refolding parameters. We identified a standard rapid dilution refolding protocol by screening different parameters including pH, temperature, composition of the refolding buffer, redox conditions and type of denaturing agents that result in a high yield of the correctly folded product with minimum loss of starting material. Although, a key to refolding is in the intermediate concentration of the denaturant in the presence of oxidizing agent, we refolded Omi without introduction of any denaturant in refolding buffer. The reason might be that the unfolded Omi are relatively soluble and can not be readily converted to a more compact structure. Thus, absence of any denaturant and presence of glycerol or sucrose enhanced protein to collapse into a compact structure as well as its stability [25]. Arginine is the most frequently used aggregation suppressor that reduces association of folding intermediates without interfering with refolding process with an unknown mechanism. Initial concentration, 40 to 50 μg/mL of the denatured Omi resulted in the highest proportion of the refolded product. In fact, protein refolding involves intermolecular interactions and follows first order kinetics and thus refolding yields commonly be decreased with increasing initial concentrations of the unfolded protein favor non-specific aggregation by a second or higher order kinetic process [26,27].
Conclusion
The method that we reported here is the first time the production of a large amount of proper biologically active recombinant Omi from bacterially expressed soluble protein aggregates and that Omi can be used for its functional, structural and protein-protein interaction analyses to evaluate the potential neuroprotective functions. In addition, this strategy can also be utilized to obtain active recombinant proteins that are expressed as soluble aggregates.
Acknowledgements
This work was supported by grants from the Next- Generation BioGreen 21 Program (PJ0079732011), Rural Development Administration of the Korean Ministry of Food, Agriculture, Forestry, and Fisheries.
References
- Clausen T, Southan C, Ehrmann M. The HtrA family of proteases: Implications for protein composition and cell fate. Mol Cell 2002; 10: 443-455.
- Spiess C, Beil A, Ehrmann MA. Temperature-depen- dent switch from chaperone to protease in a widely conserved heat shock protein. Cell 1999; 97: 339-347.
- Martins LM, Turk BE, Cowling V, Borg A, Jarrell ET, Cantley LC, Downward J. Binding specificity and regulation of the serine protease and PDZ domains of HtrA2/Omi. J Biol Chem 2003; 278: 49417-49427.
- Suzuki Y, Imai Y, Nakayama H, Takahashi K, Takio K, Takahashi R. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, induc- ing cell death. Mol Cell 2001; 8: 613-621.
- Hegde R, Srinivasula SM, Zhang Z, Wassell R, Mukat- tash R, Cilenti L, DuBois G, Lazebnik Y, Zervos AS, Fernandes-Alnemri T, Alnemri ES. Identification of Omi/HtrA2 as a mitochondrial apoptotic serine prote- ase that disrupts inhibitor of apoptosis protein-caspase interaction. J Biol Chem 2002; 277: 432-438.
- Martins LM, Morrison A, Klupsch K, Fedele V, Moisoi N, Teismann L, Abuin A, Grau E, Geppert M, Livi GP, Creasy CL, Martin A, Hargreaves I, Heales SJ, Okada H, Brandner S, Schulz JB, Mak T, Downward J. Neu- roprotective role of the reaper-related serine protease HtrA2/Omi revealed by targeted deletion in mice. Mol Cell Biol 2004; 24: 9848-9862.
- Jones JM, Datta P, Srinivasula SM, Ji W, Gupta S, Zhang Z, Davies E, Hajnoczky G, Saunders TL, Keuren MLV, Fernandes-Alnemri T, Meisler MH, Al- nemri ES. Loss of Omi mitochondrial protease activity causes the neuromuscular disorder of mnd2 mutant mice. Nature 2003; 425: 721-727.
- Gray CW, Ward RV, Karran E, Turconi S, Rowles A, Viglienghi D, Southan C, Barton A, Fantom KG, West A, Savopoulos J, Hassan NJ, Clinkenbeard H, Hanning C, Amegadzie B, Davis JB, Dingwall C, Livi GP, Creasy CL. Characterization of human HtrA2, a novel serine protease involved in the mammalian cellular stress response. Eur J Biochem 2000; 267: 5699-5710.
- Park HJ, Seong YM, Choi JY, Kang S, Rhim H. Alz-
- Tsumoto K, Abe R, Ejima D, Arakawa T. Non- denaturing solubilization of inclusion bodies. Curr Pharm Biotechnol 2010; 11: 309-312.
- Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol 2004; 22: 1399-1408.
- Ventura S, Villaverde A. Protein quality in bacterial inclusion bodies. TRENDS Biotechnol 2006; 24: 179-
- Haacke A, Fendrich G, Ramage P, Geiser M. Chaper- one over-expression in Escherichia coli: apparent in- creased yields of soluble recombinant protein kinases are due mainly to soluble aggregates. Protein Expr Pu- rif 2009; 64: 185-193.
- Martinez-Alonso M, Gonzalez-Montalban N, García- Fruitos E, Villaverde A. The functional quality of solu- ble recombinant polypeptides produced in Escherichiacoli is defined by a wide conformational spectrum. Ap- plied Environ Microbiol 2008; 74: 7431-7433.
- Martinez-Alonso M, Gonzalez-Montalban N, García- Fruitos E, Villaverde A. Learning about protein solubil- ity from bacterial inclusion bodies. Microb Cell Fact 2009; 8: 4. doi: 10.1186/1475-2859-8-4.
- Nomine Y, Ristriani T, Laurent C, Lefevre JF, Weiss E, Trave G. Formation of soluble inclusion bodies by HPV E6 oncoprotein fused to maltose-binding protein. Protein Expr Purif 2001; 23: 22-32.
- Garcia-Fruitos E, Martinez-Alonso M, Gonzalez- Montalban N, Valli M, Mattanovich D, Villaverde A. Divergent genetic control of protein solubility and con- formational quality in Escherichia coli. J Mol Biol 2007; 374: 195-205.
- Vera A, Gonzalez-Montalban N, Aris A, Villaverde A.
- Schrodel A, De Marco A. Characterization of the ag- gregates formed during recombinant protein expression in bacteria. BMC Biochem 2005; 6:10. doi: 10.1186/- 1471-2091-6-10.
- Savopoulos JW, Carter PS, Throne S, Pettman GR, Karran EH, Gray CW, Ward RV, Jenkins O, Creasy CL. Expression, purification, and functional analysis of the human serine protease HtrA2. Protein Expr Purif 2000; 19: 227-234.
- Cilenti L, Lee Y, Hess S, Srinivasula S, Park KM, Jun- queira D, Davis H, Bonventre JV, Alnemri ES, Zervos AS. Characterization of a novel and specific inhibitor for the pro-apoptotic protease Omi/HtrA2. J Biol Chem 2003; 278: 11489-11494.
- Kim DY, Kim DR, Ha SC, Lokanath NK, Lee CJ, Hwang HY, Kim KK. Crystal structure of the protease domain of a heat-shock protein HtrA from Thermotogamaritime. J Biol Chem 2003; 278: 6543-6551.
- Skorko-Gloneka J, Zurawaa D, Tanfanib F, Scireb A, Wawrzynowc A, Narkiewicza J, Bertolid E, Lipinskaa B. The N-terminal region of HtrA heat shock protease from Escherichia coli is essential for stabilization of HtrA primary structure and maintaining of its oli- gomeric structure. Biochim Biophys Acta 2003; 1649: 171-182.
- Choudhury D, Roy S, Chakrabarti C, Biswas S, Datta- gupta JK. Production and recovery of recombinant propapain with high yield. Phytochem 2009; 70: 465-
- Tsumoto K, Ejima D, Kumagai I, Arakawa T. Practical considerations in refolding proteins from inclusion bod- ies. Protein Expr Purif 2003; 28: 1–8.
- Vallejo LF, Rinas U. Strategies for the recovery of ac- tive proteins through refolding of bacterial inclusion body proteins. Microb Cell Fact 2004; 3:11.
- Ventura S, Villaverde A. Protein quality in bacterial inclusion bodies. TRENDS Biotechnol 2006; 24: 179- 185.