Research Article - Biomedical Research (2017) Volume 28, Issue 20
Isolation of Stenotrophomonas maltophilia from water and water tap
Rabeeh Izadi Amoli1*, Jamileh Nowroozi2, Azar Sabokbar3, Sadegh Fattahi4, Galia Amirbozorgi5
1Department of Microbiology, Ayatollah Amol Branch, Islamic Azad University, Amol, Iran
2Microbiology Department of Islamic Azad University, North Tehran Branch, Iran
3Department of Microbiology, Karaj Branch, Islamic Azad University, Karaj, Iran
4North Research Center Pasteur Institute of Iran, Iran
5North Research Center Pasteur Institute of Iran, Amol, Iran
- *Corresponding Author:
- Rabeeh Izadi Amoli
Department of Microbiology Ayatollah Amol Branch Islamic Azad University Iran
Accepted date: September 25, 2017
Abstract
Background and objectives: Stenotrophomonas maltophilia is one of the pathogenic bacteria originated from water and is released through water. This study was conducted with the aim of isolating Stenotrophomonas maltophilia from water and water tap in the environment and investigating the existence of rpfF gene and its relation with biofilm formation, motility and the extracellular enzymes of bacteria.
Methods: This cross sectional descriptive study was done on 150 samples of water and 100 samples of water tap in the environment. First, samples were identified through cultivation and molecular method. Existence of rpfF gene of isolates was investigated by PCR method. Formation of biofilm by microplate method and staining crystal violet was investigated. Swimming, swarming and twitching motilities of bacteria and also production of extracellular enzymes of isolates were investigated. In order to find out the relationship between rpfF gene and biofilm formation, motility and production of Extracellular enzymes, Pearson’s statistical analysis was used.
Results: The rate of Stenotrophomonas maltophilia isolation was 1.2%. 100% of isolates included rpfF gene. 100% of isolates of Stenotrophomonas maltophilia were able to move and form biofilm. Enzymes such as gelatinase, lecithinase and protease were included in 100% of isolates and hyaluronidase, lipase and hemolysin of them are included in 66.66% of isolates.
Conclusions: The results of this study suggested the existence of Stenotrophomonas maltophilia bacterium in water and water tap. Also, there was a significant relationship between the existence of rpfF gene and biofilm formation and extracellular enzymes and positive relationship with motility of bacteria.
Keywords
Stenotrophomonas maltophilia, Water, Enzymes, Motility, Biofilm.
Introduction
Stenotrophomonas maltophilia is a gram-negative and nonfermented bacterium. Growing outbreak of hospital infections originating from this bacterium is considered important in immunocompromised individuals because this pathogenic bacterium is related to the considerable cases of patients’ death [1,2].
Stenotrophomonas maltophilia is a water source organism and has multiple drug resistance which is possible to be at risk of breaking out in or out of hospital. These bacteria exist in Aquatic and marine environments with low or lack of nutritional materials. Besides, the number of Stenotrophomonas maltophilia cells in water samples, depending on environmental conditions including how to use water source and heat, is different [3].
Plumbing water can include pathogenic factors to the extent that it’s important for immunocompromised individuals [4]. This bacterium is isolated from several sources and can be found on plumbing water [5]. Sink pores, lakes and rivers, biofilms on the fractured surfaces, aqueous solutions, plumbing and bottle water, ice machines and sinks [3].
Stenotrophomonas maltophilia has flagella and fimbriae which help bacteria adhere well to the living and non-living surfaces [6]. Also, it’s able to form biofilm in systems of water distribution and be a risky factor for immunocompromised individuals [1,7]. In drainage of aquatic environment, these bacteria may be deposit and form biofilm [3]. Enzymatic activities of bacteria help them grow and survive in environment [8].
Bacterial cells are not aware of the density of other microorganisms but can indirectly identify the bacterial congestion by marker system which intervene the production of certain molecules [9] and this action is called Quorum Sensing (QS).
In contrary to gram negative bacteria, Stenotrophomonas maltophilia has a kind of cell to cell signal factor which is too much similar to the system associated to Diffusion Signal Factor (DSF) from herbal pathogenic bacteria called Xanthmonas campestris, instead of using autoinducer molecules or LuxIR [10]. Quorum sensing system in this bacterium is a regulating factor in pathogenicity of Stenotrophomonas maltophilia. Production of DSF which is essential for quorum sensing is under the control of rpfF gene [11].
This study aims to isolate Stenotrophomonas maltophilia from water and water tap and investigate the existence of rpfF gene and its relation to biofilm formation, motility and extracellular enzymes.
Materials and Methods
A hundred and fifty water and 100 water tap samples were collected in Amol of Iran in 2016. Simple sampling method was used and the number of samples were determined from the previous works. Water sampling was done in sterile condition and sterile dish. First, water flows out of tap pipe for 5 min and then the water tap was disinfected by flame and chlorinated water was neutralized by sodium thiosulfate. Then samples passed through sterile filter and filter transferred to inside of Trypticase Soy Broth (HiMedia). Sampling of water tap was done by swab, it firstly transferred to Trypticase Soy Broth (HiMedia) and in both cases, after 24 h, the process continues.
After cultivating the samples in blood agar and MacConkey agar broth, observation of gram negative bacilli, biochemical tests were done for detecting bacterium.
DNA was extracted by high pure PCR template preparation kit based on the following protocol:
Two hundred microliter culture medium including bacterium has been grown and spilled into a 1.5 μl microtube. Centrifuging was done for 5 min with round of 3000 XG. The solution on the surface was thrown away and 200 μl PBS and 5 μl lysozyme was added to the sediment and kept for 15 min under 37°C. Then, adding 200 μl of binding buffer and 40 μl of proteinase K, it was kept for 15 min under 70°C. 100 μl isopropanol was added and mixed up. The solution was transferred to the new filter. Then, it was centrifuged for 2 min and with round of 8000 XG. 500 μl of Inhibitor removal buffer was added and centrifuged for 1 min with the round of 8000 XG. 500 μl of wash buffer solution was added. Centrifuging was done for 1 min with round of 8000 XG. Two previous steps were redone. And centrifuging was done with high speed (12000 XG) for 10 s. A filter was placed in 1.5 μl-clean microtube, 200 μl elution buffer was added to it. Elution buffer got heated under 70°C before consumption. Centrifuging was done for 1 min with the round of 8000 XG. The new microtube contained DNA and the filter thrown away.
The elements of PCR reaction including 5 μL from DNA, 0.2 μL from enzyme of Taq DNA Polymerase, 1 μL from every primer (F: GCTGGATTGGTTCTAGGAAAACGC and R: ACGCAGTCACTCCTTGCG) of 23 S rRNA gene [12], 0.5 μL from dNTP, 2.5 μL from buffer, 0.75 μL from MgCl2, 14.05 μL from deionized water mixed and the final volume was 25 μl. For rpfF gene, the reaction element was the same. The primers related to this gene were designed by AllelID 6 which included R: CGAGGAAGGCGTGTTGATGG and F: CTGGCTGGCGGTGTAGAGG.
PCR reaction was done in Thermocycler instruments (TECHNE). Thermal cycler for 23 S rRNA gene included:
Initial denaturation for 4 min under 94°C, 35 times denaturation under 94°C for 30 s, annealing for 1 min under 56°C, Extension for 35 s under 72°C and final extension for 5 min under 72°C. Thermal cycle for investigation of rpfF gene included initial denaturation for 4 min under 94°C, 35 times denaturation under 94°C for 35 s, annealing for 1 min under 59°C, extension for 45 s under 72°C and final extension for 5 min under 72°C. After the reaction, the production of both genes on agarose gel 2% became electrophoresis.
In order to investigate swimming motility, the cultivated bacteria have been inoculated on LB agar in tryptone broth with agarose and incubated under 30°C for 12-14 h. In order to investigate swarming motility, the 24 h cultivated bacteria on LB agar, was cultivated point by point in nutrient broth to which agar and glucose was added and incubated for 24 h under 37°C. In order to investigate twitching motility, bacteria cultivated in LB agar were used and cultivated in LB broth with agar and incubated for 24 h under 37°C.
The results were investigated based on the amount of halo formed. The results were reported as semi-quantitative as 1+, 2+, 3+ and 4+ [8].
Gelatinase enzymes (cultivation in gelatin agar broth), hemolysis (cultivation in tryptic soy agar broth that includes 5% of sheep blood), lecithinase (cultivation in tryptic soy agar broth that includes yolk), hyaluronidase (cultivation in Muller Hinton agar reinforced by bovine serum albumin (final concentration 1%) and includes hyaluronic acid), lipase (cultivation in tryptic soy agar broth reinforced by 1% tween 80) and proteinase (cultivation in Muller Hinton agar that included 3% of skim milk) were tested [8].
In order to investigate the formation of biofilm, microplate method and crystal violet painting were used and glacial acetic acid was added to each sink and reading optical absorption in 492 nm by Elisa reader device [13]. For each sample, three repetitions occurred and samples were classified as bellow:
ODC (Optical Density Cut off)=the mean of optical absorption of control (SD × 3)
Un-formation of biofilm=(OD ≤ ODC)
Formation power of weak biofilm=ODC<OD ≤ (2 × ODC)
Formation power of moderate biofilm=(2 × ODC)<OD ≤ (4 × ODC)
Formation power of strong biofilm=(4 × ODC)<OD
Statistical analysis was done by SPSS VERGEN 16. Descriptive statistics were used to describe the basic features of the data. Pearson’s correlation test was used to evaluate the data.
Results
Three isolates of Stenotrophomonas maltophilia, 1 isolate of water tap (1%) and 2 isolates of water (1.33%) were isolated by cultivation method, biochemical tests and molecular method by targeting 23 S rRNA.
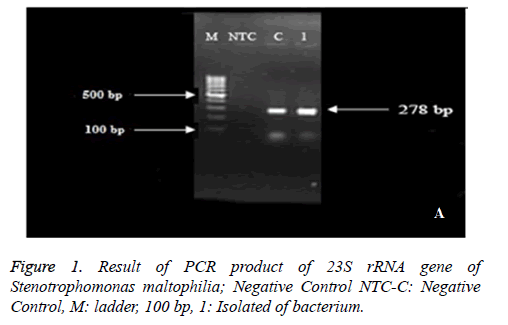
Isolates with 23 S rRNA formed band around 278 bp (Figure 1). The production of PCR was sent to Macro gene Company in South Korea in order to determine the sequence and the results of sequence determination have approved the given bacteria after blasting in NCBI.
One isolate from water was recorded in NCBI site with Accession Number: KX813715 under the name of Stenotrophomonas maltophilia strain RS 23 S ribosomal RNA gene, partial sequence.
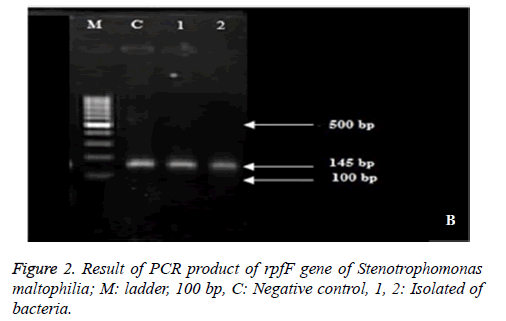
Result of PCR product of rpfF gene of Stenotrophomonas maltophilia suggested that 100 % of isolates included rpfF gene and formed band around 145 bp (Figure 2).
100% of isolates of Stenotrophomonas maltophilia had motility. A hundred percent of isolates had strong swimming motility. There has been a strong swarming motility in 66.66% of isolates (2 isolates from water) and weak swarming motility in 33.33% of isolates (1 isolate of water tap). Also there has been a Strong twitching motility in 66.66% of isolates (2 isolates from water) and weak twitching motility in 33.33% of isolates (1 isolate from water tap). As the variance analysis shows, there is a significant difference between strong and weak motility of bacteria (P<0.0).
The results of Pearson’s correlation test between the type and the level of motility of bacteria tested by control bacteria with rpfF gene suggested that the correlation coefficient between rpfF gene and type and level of bacteria motility was positive and weak and was not statistically significant (P>0.05).
Formation of biofilm was seen in 100% of isolates, 33.33% of isolates were powerful to form weak biofilm, 66.66% of isolates were powerful to form moderate biofilm and none of them were powerful to form strong biofilm.
Also, the results of correlation between the existence of rpfF gene and biofilm formation in tested bacteria and control bacteria suggested that there is a positive, strong and significant correlation between these two variables (P<0.05).
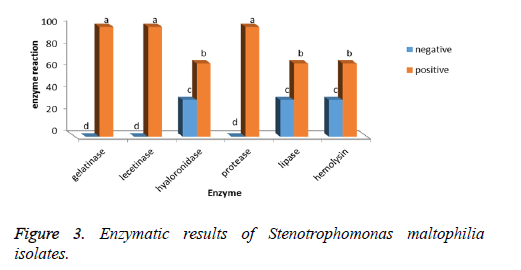
Gelatinase, lecithinase, and protease enzymes are found in 100% of Stenotrophomonas maltophilia isolates and hyaluronidase, lipase and hemolysin enzymes are found in 66.66% of bacteria isolates (Figure 3).
The investigation of Pearson’s correlation between the existence of rpfF gene and existence of enzymes such as gelatinase, lystinase, hyaluronidase, protease, lipase and hemolysin suggested that there is a positive, strong and significant correlation between these two variables (98%).
Discussion
In this article, Stenotrophomonas maltophilia was isolated from water and water tap. Stenotrophomonas maltophilia naturally resides in water. A climate change in spreading of the infection resulting from Stenotrophomonas maltophilia through transference of infections, factors resulting from water is effective and that’s why the safety of drinking water tanks is important [3]. Temperature changes causes geographical outbreak of diseases from water. Changes in rainfall and rain and also changes in chemical composition, due to the pollution, can change the variation of communities of existing microbes and these changes will result in emerging the new pathogenic bacteria [14].
Increasing the global warming, most probably causes growing of cells and if the cells are in close contact with sensitive people, they will be active and bring about the risk of increasing infections. Also, increasing the cellular density of these bacteria in aqueous environments may result in entering external DNA and receiving following important drugresistance and pathogenic genes. Entering the bacteria through water to clinical resources seems logical.
Controlling the infection of this resource can help to control these bacteria in clinical units. So, physicians and clinics must consider water and water tap as a potential source not only for Stenotrophomonas maltophilia bacterium, but also for other bacteria such as Pseudomonas aeruginosa, Acinetobacter baumanii, and Legionella pneumophila [15]. When Stenotrophomonas maltophilia accompany other bacteria of MDR (Multi-Drug-Resistance) including Klebsiella, Serratia, Sphingomonas and Citrobacter, there will be problems relating to transference of gene in microbe communities of environments such as sewage and biofilm existing in water pipe system by receiving DNA from other bacteria species. It was reported that Stenotrophomonas maltophilia transfers antibiotic resistance genes to other bacteria including Pseudomonas aeruginosa and family member of Enterobacteriaceae [3].
In present study, Stenotrophomonas maltophilia bacterium has been isolated from 1% of water tap samples and 1.33% of water samples. In the study of Adjide et al. in 2010, Stenotrophomonas maltophilia bacterium has been found in 2.5% of pipe water swab which was higher than what was found in this study [16], luckily, the present study suggested less infection.
In the present study, all of the samples had motility, 100% of isolates had strong swimming motility, 66.66% had strong swarming motility and 33.33% had weak twitching motility. 66.66% of isolates had strong twitching motility and 33.33% of them had weak twitching motility. In a study done by Pompilio et al. out of 89 isolates of Stenotrophomonas maltophilia [4], non-clinical samples did not have any motility, of course the type of motility being investigated was swimming and twitching [17] but in the present study, all of the isolates of bacteria had motility. One of the reasons of bacteria motility is food reception because in these environments, contrary to the clinical ones, there may be low level of food. It is concluded that different motility behaviors are seen in the given bacteria.
There may be a relationship between the type of motility and the environment that bacteria reside. Some of the bacteria have two flagella used for motility in different environmental conditions [18].
All of the bacteria isolates had enzymes of gelatinase, lecithinase and protease but lipase, hemolysin, hyaluronidase enzymes included in 66.66% of isolates. It is likely that these enzymes in clinical isolates, due to red blood cells, lipids and hyaluronic acid of host tissue, are high. Enzymatic activities of bacteria, helps the pathogenicity of bacteria and clinical isolates are able to get the genes producing these enzymes from outside of the hospital.
One of the other factors which help the pathogenicity of bacteria is biofilm formation. In this study, all of the isolates were able to form biofilm, but some of them were able to form moderate biofilm and the others to form weak ones. In a study done by Bonoventura et al. it was concluded that the ability of forming biofilm is effected by different environmental conditions such as heat, access to oxygen and pH [19].
One of the adjusting factors of pathogenicity of Stenotrophomonas maltophilia bacteria is rpfF gene [11]. This gene plays role in adjusting the motility, forming biofilm and producing protease extracellular enzymes and antibiotic resistance of bacteria. Mutant strains of rpfF gene suggested the decrease in ability of movement and biofilm formation. Adjusting the formation of biofilm depends on rpfF gene [17]. In this study, all of the isolated bacteria had rpfF gene. In the study of Zhuo et al. the frequency of rpfF gene in Stenotrophomonas maltophilia bacteria was 50% [20] and in the study of Pompillio et al., it was 61.8% [17].
The results of the studies of Zhuo et al. and Pompillio et al. were different from those of this study in terms of the frequency of rpfF gene. So this gene is not seen in all isolates of bacteria. The results of this study suggest that all of the isolates could form biofilm, had motility, especially strong swimming motility and also gelatinase, lecithinase and protease enzymes were seen in all isolates, so we can conclude that this gene is related to this pathogenic factors., as Pompillio et al. but in terms of gelatinase enzymes, lecithinase was not mentioned [17].
The results of this study, suggested the existence of Stenotrophomonas maltophilia bacterium in water and water tap. Also, there was a significant relationship between the existence of rpfF gene with biofilm formation and extracellular enzymes and also a positive relationship with the type of bacteria motility.
Acknowledgements
We are grateful to the molecular lab staff from the Pasteur Institute of Iran (North research Center) and Islamic Azad University Ayatollah Amoli Branch for their sincere cooperation.
References
- Brooke JS. Biofilm production of clinical isolates of Stenotrophomonas maltophilia altered by sodium phosphate buffer supplementation of the medium. Ann Microbiol 2007; 57: 677.
- Senol E. Stenotrophomonas maltophilia: the significance and role as a nosocomial pathogen. J Hosp Infect 2004; 57: 1-7.
- Brooke JS. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev 2012; 25: 2-41.
- Feazel LM, Baumgartner LK, Peterson KL, Frank DN, Harris JK, Pace NR. Opportunistic pathogens enriched in showerhead biofilms. Proc Natl Acad Sci USA 2009; 106: 16393-16399.
- Sakhnini E, Weissmann A, Oren I. Fulminant Stenotrophomonas maltophilia soft tissue infection in immunocompromised patients: an outbreak transmitted via tap water. Am J Med Sci 2002; 323: 269-272.
- Pompilio A, Crocetta V, Confalone P. Adhesion to and biofilm formation on IB3-1 bronchial cells by Stenotrophomonas maltophilia isolates from cystic fibrosis patients. BMC Microbiol 2010; 10: 102.
- Sacchetti R, De Luca G, Zanetti F. Control of Pseudomonas aeruginosa and Stenotrophomonas maltophilia contamination of microfiltered water dispensers with peracetic acid and hydrogen peroxide. Int J Food Microbiol 2009; 132: 162-166.
- Thomas R, Hamat RA, Neela V. Extracellular enzyme profiling of Stenotrophomonas maltophilia clinical isolates. Virulence 2014; 5: 326-330.
- Bjarnsholt T. The role of bacterial biofilms in chronic infections. APMIS 2013; 136: 1-51.
- Fouhy Y, Scanlon K, Schouest K. Diffusible signal factor-dependent cell-cell signaling and virulence in the nosocomial pathogen Stenotrophomonas maltophilia. J Bacteriol 2007; 189: 4964-4968.
- Huedo P, Yero D, Martinez-Servat S. Two different rpf clusters distributed among a population of Stenotrophomonas maltophilia clinical strains display differential diffusible signal factor production and virulence regulation. J Bacteriol 2014; 196: 2431-2442.
- Gallo SW, Ramos PL, Ferreira CA, Oliveira SD. A specific polymerase chain reaction method to identify Stenotrophomonas maltophilia. Mem Inst Oswaldo Cruz 2013; 108.
- Fonseca AP, Extremina C, Fonseca AF, Sousa JC. Effect of subinhibitory concentration of piperacillin/tazobactam on Pseudomonas aeruginosa. J Med Microbiol 2004; 53: 903-910.
- Shuman EK. Global climate change and infectious diseases. N Engl J Med 2010; 362: 1061-1063.
- Safdar A, Rolston KV. Stenotrophomonas maltophilia: changing spectrum of a serious bacterial pathogen in patients with cancer. Clin Infect Dis 2007; 45: 1602-1609.
- Adjide CC, De Meyer A, Weyer M. Stenotrophomonas maltophilia and Pseudomonas aeruginosa water-associated microbiologic risk assessment in Amiens University Hospital Centre. Pathol Biol (Paris) 2010; 58: 1-5.
- Pompilio A, Pomponio S, Crocetta V. Phenotypic and genotypic characterization of Stenotrophomonas maltophilia isolates from patients with cystic fibrosis: genome diversity, biofilm formation, and virulence. BMC Microbiol 2011; 11: 159.
- Eisenbach M. Bacterial chemotaxis. Encyclopedia of life sciences. Nature Publishing Group 2001.
- Di Bonaventura G, Stepanovic S, Picciani C, Pompilio A, Piccolomini R. Effect of environmental factors on biofilm formation by clinical Stenotrophomonas maltophilia isolates. Folia Microbiol (Praha) 2007; 52: 86-90.
- Zhuo C, Zhao QY, Xiao SN. The impact of spgM, rpfF, rmlA gene distribution on biofilm formation in Stenotrophomonas maltophilia. PLoS One 2014; 9: 108409.