Research Article - Journal of Fisheries Research (2018) Volume 2, Issue 1
Isolation and identification of Citrobacter spp. from the intestine of Procambarus clarkii.
Honglian Chen, Yongjie Wang*, Jing Zhang, Yunsheng Chen, Minglin WuFisheries Research Institute, Anhui Academy of Agricultural Sciences, Hefei, China.
- *Corresponding Author:
- Yongjie Wang
Fisheries Research Institute
Anhui Academy of Agricultural Sciences
Hefei
China
Tel: +861082106755
E-mail: hfwangyongjie@163.com
Accepted date: April 09, 2018
Citation: Chen H, Wang Y, Zhang J, et al. Isolation and identification of Citrobacter spp. from the intestine of Procambarus clarkii. J Fish Res. 2017;2(1):1-6.
DOI: 10.35841/fisheries-research.2.1.1-6
Visit for more related articles at Journal of Fisheries ResearchAbstract
Citrobacter have caused a wide spectrum of infectious diseases in aquaculture animals and human. In previous study, pathogenic Citrobacter spp. was frequently isolated from hepatopancreas of diseased Procambarus clarkii. It suggested that the intestine might be the reservoirs of Citrobacter spp. Intestinal flora were isolated from P. clarkii with Salmonella shigella agar. 126 isolates were performed with enterobacterial repetitive intergenic consensus (ERIC) PCR. 95 strains of seven ERIC-PCR electrophoretic genotypes were identified as Citrobacter spp. by 16S rRNA gene sequence analysis and the urease gene detection of the representative strains. Eight representative strains were defined by biochemical characteristics and multilocus sequence analysis of genes fusA, leuS, rpoB and pyrG. Isolates 425C1, 731C1 and 519C4 were determined as C. freundii, C. werkmanii and a new species of C. freundii complex, respectively. Isolates 425C3, 519C1, 519C3, 519C41 and 519C53 were identified as C. braakii. C. freundii strain 425C1 and C. werkmanii strain 731C1 were verified pathogenic bacteria to P. clarkii and Kunming mice in varying degrees. To our knowledge, this is the first report of pathogenic C. werkmanii.
Keywords
Citrobacter, Procambarus clarkii, ERIC-PCR, 16S rRNA, Multilocus sequence analysis
Introduction
Citrobacter species were considered the intestinal inhabitants of human and animals, and commonly existed in sewage, water and soil [1]. There are 12 recognized species within the genus Citrobacter[2], of which three were pathogenic bacteria in human. C. farmeri andC. koseri hadcaused meningitis in immunocompetent patient [3,4]. C. freundii wasan importantfood-borne and hospital-acquired pathogen, which had caused diarrhea, urinary tract infection, peritonitis, bacteremia, brain abscess, and meningitis [5-9]. C. freundii was now generally considered an opportunistic pathogen in aquaculture. It has associated with a wide spectrum of infectious diseases in aquaculture animals, involving gastroenteritis of one-year rainbow troutOncorhynchus mykiss [10], red-leg syndrome of Rana catesbeiana [11], septicaemia of Garra rufa obtusaand Ziphius carvirostris [12,13], systemic infection with no typical syndrome of Carcharhinus melanopterus [14], hepatopancreas necrosis and rotted gill of Cherax quadricarinatus, Portunus trituberculatus and Procambarus clarkii [15-17]. C. braakii had caused septicaemia of Crocodile niloticus [18].C. gillenii and Citrobacter spp. had caused disease of O. mykiss [20].
The red swamp crayfish C. freundii, native to the southeastern United States, has been successfully exploited as a fishery product and cultured worldwide. It is an important economic species in inland China due to its large-scale processing and exporting and its gradual development of domestic market value. Recently, white spot syndrome (WSS) causes high mortality and large economic losses of cultured P. clarkii in inland China. Pathogenic Citrobacter spp. were frequently isolated from hepatopancreas of diseased P. clarkii affected WSSV. C. freundii from the intestine of C. idellus has been proved having the pathogenicity to the wild zebrafish and Kunming mice [20]. That causes our interesting to investigate Citrobacter spp. in the intestine of crayfish. The objective of this study was to isolate Citrobacter bacteria from the intestine of P. clarkii,clarify taxonomy of the representative strains, and describe their potential pathogenicity and antibiotics susceptibility.
Materials and Methods
Isolation of the intestinal flora from P. clarkii
Intestinal flora was isolated from twenty seemingly healthy P. clarkii (weight ca. 20 g per crayfish), of which ten crayfishes were purchased from supermarket, others were collected from a crayfish farm in Hefei, China. 0.5 g intestine was took and made into homogenate, then ten fold serial diluted, 0.1 mL diluent was coated onto Salmonella shigella (SS) agar plate (Rishui, Qingdao, China) and incubated at 28°C for 24 h. A total of 126 isolates were picked up according to the proportion of each color colonies on SS agar plate, purified, and used for enterobacterial repetitive intergenic consensus (ERIC) PCR genotyping analysis.
DNA extraction, ERIC-PCR analysis, 16S rRNAgene sequence analysis and urease gene detection
Bacterial isolates were cultured in brain heart infusion broth. DNA was extracted using ezup bacterial genomic DNA extraction kit (TIAGEN). ERIC-PCR was performed using 25 μL PCR mix, with primers ERIC1: 5’-ATGTAAGCTCCTGGGGATTCAC-3’ and ERIC2: 5’-AAGTAAGTGACTGGGGTGAGCG-3’. The amplification was performed using the following conditions: initial denaturation for 4 min at 94°C, 35 cycles of 94°C for 45 s, 52°C for 45 s and 72°C for 2.5 min, then 10 min at 72°C. PCR products were visualised by gel electrophoresis on a 1% agarose gel. One isolate was chose from each genotype as the representative strain. The extracted DNAs of the representative strains were used for PCR amplification of the partial 16S ribosomal RNA sequence using universal primer pairs (27F, 1492R). Primers and PCR conditions were the same as those described by Lü et al. [20]. Positive PCR products were sent to Sangon Biotech for sequencing. The achieved sequences were carried out by BLAST retrieval system in NCBI. All isolates of the ERIC-PCR genotypes with their representative strains identified as Citrobacter spp., were detected the Citrobacterurease gene with the primers F1: 5’-TGAAGCTGAACTACCCGGAATC-3’ and R1: 5’-TGTCCAGGCTCAAAACGTAC-3’, PCR was carried out using the conditions: 4 min at 94°C, 33 cycles of 40 s at 94°C, 40 s at 55°C and 1 min at 72°C, then 7 min at 72°C. PCR products were visualised by gel electrophoresis on a 2% agarose gel and the expected PCR product was 454 bp.
Biochemical and molecular characterization
The gram staining test was performed using the Hucker method. The biochemical characteristics of the representative Citrobacterstrains were performed using commercial microtest systems (Hangzhou Binhe Microorganism Reagent Co., Ltd, China).
The extracted DNAs of the representative Citrobacter strains were used for PCR amplification of the β subunit of RNA polymerase gene rpoB, the CTP synthetase gene pyrG, the protein synthesis elongation factor-G gene fusA, and the leucine tRNA synthetase gene leuS. Primers and PCR conditions were the same as those described by Delétoile et al. [21]. The PCR products were purified by TaKaRa MINIBEST agarose gel DNA extraction kit, cloned into the pMD19-T vector (TaKaRa) and transformed into competent E. coli DH5a. Positive clones were sent to Sangon Biotech for sequencing.
The genus Citrobacter includes 12 species: group I, also named C. freundii complex (C. freundii, C. youngae, C. braakii, C. werkmanii, C. gillenii, C. murliniae and C. pasteurii), group II (C. amaloniticus, C. farmeri, C. sedlackii and C. rodentium) and group III (C. koseri) [22-24]. Their 16S rRNA, rpoB, fusA, pyrG and leuS gene sequences were obtained from GenBank database. RpoB, fusA, pyrG and leuS gene sequences of 12 Citrobacter species and the representative strains were concatenated into a 2,082 bp alignment used for nucleotide similarity analysis and phylogenetic analysis. Similarity analysis was performed using DNAMAN 6.0 software. The 16S rRNA sequences and concatenated sequences were aligned by Clustal X2 software, then used to construct phylogenetic trees by neighbor joining method of MEGA 5.1 software with 1000 times of bootstrap analysis, respectively.
Pathogenicity assays in P. clarkii and mice
The newly cultured representative strains were adjusted to the concentration of 108 CFU/mL. Ten seeming healthy P. clarkii (ca. 20 g per crayfish) were prepared for each representative strain. Each crayfish was intramuscular injected with 0.05 mL diluent into the third abdominal segments, then cultured in aerated tap water at (25 ± 1)°C. Ten Kunming mice weighting ca. 22 g (provided by Anhui Medical University) were prepared for each isolate, each mouse was inoculated intraperitoneally with 0.5 mL diluent. Control animals received PBS alone and the observed time post-infection was 14 days. Morbid crayfish and mice were subjected to laboratory, their liver, kidney and spleen were analyzed for isolation for bacterial.
Antibiotics susceptibility test
Disc diffusion method was adopted [25]. Fresh cultured representative strains were selected and dissolved in 0.65% normal saline. The concentration was adjusted to 108 CFU/mL. The strains were evenly coated to MH agar plate by cotton swab. A total of 21 kinds of antibiotics disks were foxed upon the surface of culture medium. After cultivated at 28°C for 24 h, diameter of bacteriostasis circle was observed and recorded (including sensitive tablet).
Results
Characterization of the Citrobacter spp.
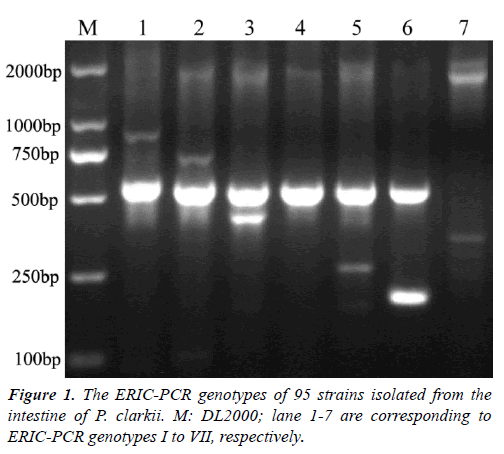
Colonies on SS agar from the intestine of P. clarkii wereround, smooth, slightly convex, entire edge and 1-2 mm in diameter, showed four different colors after 24 h incubation at 28°C. A total of 126 colonies were characterized, including 84 red colonies, 28 pink colonies, 10 pink colonies with a dark center, 4 white colonies with a dark center. The ERIC-PCRs generated a total of 13 genotypes (data not shown). According to 16S rRNA sequences analysis, the representative strains of seven ERIC genotypes shared nearest relative to genus Citrobacter. ERIC genotypes I to VI had a 500 bp band(Figure 1), genotypes I to VII contained 11, 15, 1, 10, 32, 10, 16strains, respectively. On SS agar, all strains in genotype VI displayed pink colonies with a dark center, strain 731C1 in genotype VII displayed a white colony with a dark center, other strains formed red colonies. Urease gene detection of the 95 isolates contained in genotypes I to VII were all positive with a 454 bp band (data not shown), that verified them as Citrobacterspp. The representative strains for genotypes I to VI were 519C4, 425C1, 519C53, 425C3, 519C3, 519C41, respectively, two representative strains for genotype VII were 731C1 and 519C1, because their colonies showed different colors on SS agar.
Biochemicalcharacteristics of the representative Citrobacter strains
The eight isolates were gram-negative, motive, short rods. The comparison of biochemical characteristics of the eight strains with those of some Citrobacter species in Bergey's Manual was shown in (Table 1). Biochemical characteristics of strain 425C1 were identical to those described for C. freundii, and strains 425C3, 519C1, 519C3, 519C41, 519C53 share the identical biochemical characteristics with C. braakii. Strain 731C1 shared the common biochemical characteristics with C. werkmanii. Strain 519C4 differed from C. freundii bybeingpositive for ornithine decarboxylase.
| Characteristics | 425C1 | 519C4 | CF | 425C3 | 519C1 | 519C3 | 519C41 | 519C53 | CB | 731C1 | CW |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Oxidase | - | - | - | - | - | - | - | - | - | - | - |
| O-F test | F | F | F | F | F | F | F | F | F | F | F |
| Indole | - | - | d | - | - | - | - | - | d | - | - |
| Citrate (Simmons) | + | + | d | + | + | + | + | + | d | + | + |
| H2S production | + | + | d | + | + | + | + | + | d | + | + |
| Ornithine | - | + | - | + | + | + | + | + | +a | + | - |
| KCN growth | + | + | d | + | + | + | + | + | + | + | + |
| Malonate | - | - | d | - | - | - | - | - | - | + | + |
| Sucrose | + | + | d | + | - | - | - | - | -b | - | - |
| Melibiose | + | + | + | + | + | + | + | + | d | + | - |
| Raffinose | + | + | d | - | - | - | - | - | - | - | - |
| Dulcitol | - | - | d | + | - | - | - | + | d | - | - |
| Adonitol | - | - | - | - | - | - | - | - | - | - | - |
| Arabitol | - | - | - | - | - | - | - | - | - | - | - |
| Arabinose | + | + | + | + | + | + | + | + | + | + | + |
| ONPG | + | + | d | + | + | + | + | + | d | + | + |
| Phenylalanine deaminase | - | - | - | - | - | - | - | - | - | - | - |
| Lysine decarboxylase | - | - | - | - | - | - | - | - | - | - | - |
| Esculin hydrolysis | - | - | - | - | - | - | - | - | - | - | - |
| Lactose | + | + | d | + | + | + | + | + | d | - | d |
| Cellobiose | + | - | d | + | - | + | + | - | d | - | d |
| Maltose | + | + | + | + | + | + | + | + | + | + | + |
| Xylose | + | + | d | + | + | + | + | + | + | + | + |
| Voges?Proskauer | - | - | - | - | - | - | - | - | - | - | - |
| Urea | + | + | d | + | + | + | + | + | d | + | + |
| Arginine dihydrolase | + | + | d | + | + | + | + | + | d | + | + |
| DNase | - | - | - | - | - | - | - | - | - | - | - |
| Gelatin hydrolysis | - | - | - | - | - | - | - | - | - | - | - |
| Inositol | - | - | - | - | - | - | - | - | - | - | - |
| Mannitol | + | + | + | + | + | + | + | + | + | + | + |
| Glucose, acid | + | + | + | + | + | + | + | + | + | + | + |
| Glucose, gas | + | + | d | + | + | + | + | + | d | + | + |
| Salicin | - | - | - | - | - | - | - | - | - | - | - |
| Sorbitol | + | + | + | + | + | + | + | + | + | + | d |
| Rhamnose | + | + | + | + | + | + | + | + | + | + | + |
| Nitrate reduction | + | + | + | + | + | + | + | + | + | + | + |
| Catalase | + | + | + | + | + | + | + | + | + | + | + |
| Starch | - | + | d | - | - | - | - | - | - | d |
Table 1. Biochemicalcharacteristics of the representative Citrobacter strains from P. clarkia.
Multilocus sequence analysis of the representative Citrobacter strains
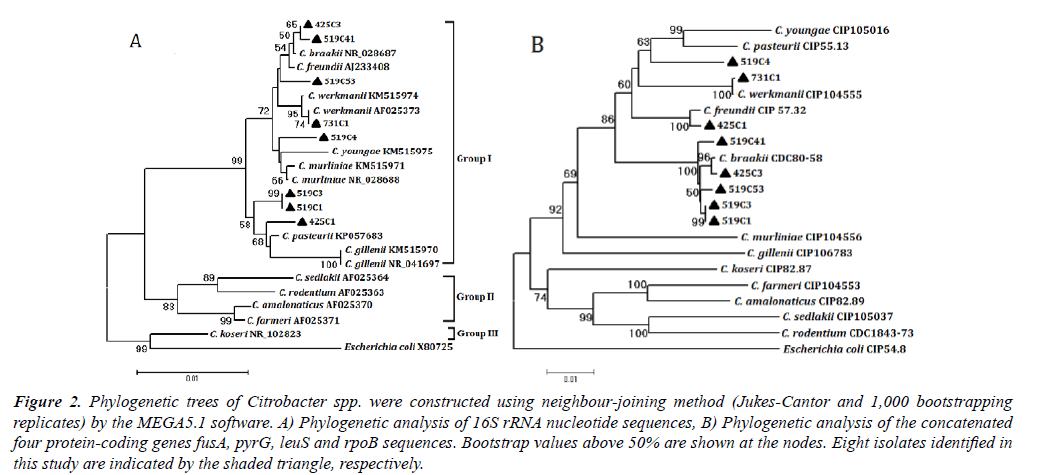
The gene sequences of the representative Citrobacterstrains isolated from the intestine of P. clarkii in this study were deposited in GenBank and available under the accession numbers KT764981 to KT764988 for 16S rRNA, KT764989 to KT764996 for fusA, KT764997 to KT765004 for leuS, KT765005 to KT765012 for pyrG and KT765013 to KT765020 for rpoB. Phylogenetic analysis of 16S rRNA sequences differentiated the eight representative strains to Citrobacter group I as defined previously [22]. Concatenated sequences of 731C1 and C. werkmanii CIP 104555 shared 99.95% similarity (1 differences in leuS gene) and formed a clade in the phylogenetic analysis (Figure 2). Concatenated sequences of 425C1 and C. freundii CIP 57.32 shared 99.57% similarity (9 differences) and formed a clade with 100% bootstrap support. Similarity values of concatenated sequence values were 99.37-99.71% between 425C3, 519C1, 519C3, 519C41, 519C53 and C. braakii and the six strains formed a clade. Similarity values of concatenated sequences ranged from 91.39% to 96.89% between strain 519C4 and 12 Citrobacter species. 519C4 was clustered in group I. Individual gene trees of the four genes confirmed these results (data not shown).
Figure 2.Phylogenetic trees of Citrobacter spp. were constructed using neighbour-joining method (Jukes-Cantor and 1,000 bootstrapping replicates) by the MEGA5.1 software. A) Phylogenetic analysis of 16S rRNA nucleotide sequences, B) Phylogenetic analysis of the concatenated four protein-coding genes fusA, pyrG, leuS and rpoB sequences. Bootstrap values above 50% are shown at the nodes. Eight isolates identified in this study are indicated by the shaded triangle, respectively.
Pathogenicity assay
The results indicated that strains 731C1 and 425C1 had the pathogenicity to P. clarkii and Kunming mice in varying extent, whereas other six isolates showed limited pathogenic to the two animals. The mortality rate of P. clarkii and Kunming mice both were 100% after intramuscular and intraperitoneally inoculated with strain 731C1. The mortality rate of P. clarkii and Kunming mice were 60% and 20% after inoculated with strain 425C1. Bacteria re-isolated from the liver, kidney and spleen of experimented moribund and dead animals were identified to be the inoculated strains 731C1 and 425C1 by biochemical characteristics and molecular biological identification,respectively.
Discussion
As is known, that Citrobacter species are the normal intestinal inhabitants of animals and human. Five Citrobacter species had been isolated from the intestine of fish. C. freundii, C. amalonaticus and C. braakii from farmed catfish were identified by biochemical characteristics [26]. C. freundii, C. gillenii and C. werkmanii from C. idell were defined by biochemical characteristics and 16S rRNA gene sequence analysis [20]. Till now, no research described Citrobacter spp. in the intestine of crayfish. In this research, C. freundii, C. braakii, C. werkmaniiand a new species of C. freundii complex were defined by biochemical characteristics and multilocus sequence analysis.
The primary presumptive identification of Citrobacterspp.fromthe intestine of P. clarkii was achieved using SS agar and ERIC-PCR. Expect that strain 731C1 displayed white colony with a dark center, other Citrobacter strains displayedred colonies, or pink colonies with a dark center on SS agar. ERIC-PCR has enough discriminatory power, and used for the molecular characterization and epidemiology investigation of some pathogens, such as Salmonella typhi, Escherichia coli, Haemophilus parasuis, Vibrio cholerae [27-30]. In this study, 95isolates of seven ERIC-PCR genotypes were verified as Citrobacterspp. by detecting the urease gene of Citrobacterspp. The fact that the typical colors of colonies on SS agar and the distinctive electrophoretic genotypes of ERIC-PCR obtained in this study suggested that the two methods would be useful tools for isolating Citrobacter spp. from complex environment. To our knowledge, this is the first report to applied ERIC-PCR technique in molecular characterization of Citrobacter spp.
The representative Citrobacter strains were distinguished to C. freundii complex (group I) by 16S rRNA gene sequence analysis. 16S rRNA gene sequence analysis could only distinguished 12 Citrobacterspecies to 3 groups, and biochemical profiles has limited capability to discriminate some strains of C. freundii complex [22-24] So multilocus sequence analysis of fusA, leuS, rpoB and pyrG genes was the critical technique to discriminate the eight strains, because the technique was proved to demonstrate limited intra-specific genetic variation and clear species demarcation of Citrobacter species and had successfully defined new species C. pasteurii [24]. Strain 425C1 was defined as C. freundii, strains 425C3, 519C1, 519C3, 519C41 and 519C53 were all identified as C. braakii and strain 731C1 was defined as C. werkmanii, according to their biochemical profiles and multilocus sequence analysis. Strain 519C4 shared the same biochemical characteristics with C. freundii except forpositive ornithine decarboxylase, and its concatenated sequences shared the highest similarity of 96.89% with that of C. freundii, while strain 519C4 did not clustered with C. freundii in phylogenetic analysis, so 519C4 was proposed a new species of C. freundii complex.
C. freundii strain 425C1 and C. werkmanii strain 731C1 have pathogenicity to P. clarkii and Kunming mice in varying degrees. C. freundii had been found to be the cause of variety diseases in aquaculture animals (see the introduction of this article). C. braakii had caused septicaemia of C. niloticus [18].C. gillenii and Citrobacter sp. had caused disease of O. mykiss [19]. This is the first report of pathogenic C. Werkmanii.
Aeromonas hydrophila, V. parahaemolyticus, WSSV, spiroplasma and C. freundii had been reported caused various diseases of P. clarkii [31-35]. Recent years, WSS was the most prevalent disease of P. clarkii in inland China and Citrobacter spp. were the commonly opportunistic pathogen co-infected with WSSV in our previous study (data not shown). The 75.40 percent isolates of the microbe on SS agar from P. clarkii were Citrobacter spp. It revealed that the intestine of C. freundii served as a reservoir of Citrobacter spp. Control pathogenic Citrobacter might be helpful to reduce the mortality of P. clarkii caused by WSS.
The eight representative Citrobacterstrains were all sensitive to doxycycline, quinolone antibiotics (enoxacin, ofloxacin, fleroxacin, enrofloxacin, lomefloxacin, nalidixic acid, norfloxacin) and aminoglycoside antibiotics (neomycin, tobramycin, kanamycin, gentamicin, streptomycin) (data not shown), the results were similar to those found with C. freundii from hepatopancreas ofdiseased P. clarkii [17].These antimicrobial agents might be used to control diseases of crayfish caused by Citrobacter population.
Conclusion
C.freundii had frequently caused infectious diseases of aquaculture animals. Here, a large number of Citrobacter strains were isolated from the intestine of seemingly healthy crayfish with SS agar in central eastern China and the representative strains of seven Citrobacter ERIC-PCR genotypes were subsequently identified as C. freundii, C. braakii, C. werkmanii and a new species of C. freundii complex by means of biochemical characteristics and multilocus sequence analysis. Two representative strains, C. freundii 425C1 and C. werkmanii731C1, were confirmed as pathogens of P. clarkii and mice by challenge experiments. Furthermore, the techniques of SS agar and ERIC-PCR would be useful tools for isolating Citrobacter spp. from complex environment.
Acknowledgement
This study was supported by Science and Technology Innovative Research Team of Anhui Academy of Agricultural Sciences (18C0513), earmarked Fund for Anhui Fishery Research System (2016-84).
References
- Frederiksen W, Brenner DJ, Krieg NR, et al. Bergey's manual of systematic bacteriology, 2nd edn. Proteobacteria. 2005;2:651-656.
- Clermont D, Motreff L, Passet V, et al. Multilocus sequence analysis of the genus Citrobacter and description of Citrobacter pasteurii sp. Nov. Int J Syst Evol Microbiol. 2015;65(5):1486-90.
- Tan CK, Lai CC, Lin SH, et al. Fatal Citrobacter farmeri meningitis in a patient with nasopharyngeal cancer. J Clin Microbiol. 2010;48(4):1499-500.
- Marecos CV, Ferreira M, Ferreira MM, et al. Sepsis, meningitis and cerebral abscesses caused by Citrobacter koseri. BMJ Case Rep. 2012;50:1333-7.
- Pardia SN, Verma IC, Deb M, et al. An outbreak of diarrhea due to Citrobacter freundii in a neonatal special care nursery. Indian J Pediatr. 1980;47(1):81-4.
- Kim BN, Woo JH, Ryu J, et al. Resistance to extended-spectrum cephalosporins and mortality in patients with Citrobacter freundii bacteremia. Infection. 2003;31(4):202-7.
- Erkan D, Zeki Y, Itir Y. Citrobacter freundii peritonitis and tunnel infection in a patient on continuous ambulatory peritoneal dialysis. J Med Microbiol. 2007;57(1):125-127.
- Plakkal N, Soraisham AS, Amin H. Citrobacter freundii brain abscess in a preterm infant: A case report and literature review. Pediatr Neonatol. 2013;54(2):137-40.
- Park S, Song SH, Lee C, et al. Bacterial pathogens in first febrile urinary tract infection affect breakthrough infections in infants with vesicoureteral reflux treated with prophylactic antibiotics. Urology. 2013;81(6):1342-1345.
- Svetlana J, Dobrila J, Lj V. Citrobacter freundii as a cause of disease in fish. 2003;53(5-6):399.
- Pasteris SE, Guidoli MG, Otero MC, et al. In vitro inhibition of Citrobacter freundii, a red-leg syndrome associated pathogen in raniculture, by indigenous Lactococcus lactis CRL 1584. 2011;151(3):336-44.
- GunWook B, JiHyung K, Choresca JC, et al. Mass mortality of doctor fish (Garra rufa obtusa) caused by Citrobacter freundii infection. J Vet Clin. 2009;26(2):150-4.
- Fernandez A, Vela AL, Andrada M, et al. Citrobacter freundii septicemia in a stranded newborn Cuvier?s beaked whale (Ziphius cavirostris). J Wildl Dis. 2011;47(4):1043-6.
- Lu J, Sun J, Dong S, et al. Isolation and identification of pathogenic bacterium Citrobacter freundii from black reef shark Carcharhinus melanopterus. Trans Oceanol Limnol. 2015;4:83-8.
- Shen J, Gu Z, Pan X, et al. Isolation and identification of Citrobacter freundii from Cherax quadricarinatus. J Fish Sci China. 2005;12(2):197-200.
- Yan B, Zhang X, Liang L, et al. Detection of cfa gene and isolation and identification of the pathogen Citrobacter freundii isolated from Portunus trituberculatus. J Fish Sci China. 2012;36(3):391-8.
- Chen H, Song G, He J, et al. Isolation, identification and antibiotics susceptibility test of Citrobacter freundii from Procambarus clarkii. Anim Husb Feed Sci. 2014;6(5):238-41.
- Rosell J, Barragán A, Carbonell MD, et al. Septicaemia caused by Citrobacter braakii infection in a captive Nile crocodile (Crocodylus niloticus). J Comp Pathol. 2017;156(1):105.
- Duman U, Saticioglu IB, Buyukekiz AG, et al. Molecular characterization and antimicrobial resistance profile of atypical Citrobacter gillenii and Citrobacter sp. isolated from diseased rainbow trout (Oncorhynchus mykiss). J Glob Antimicrob Resist. 2017;10:136-42.
- Lü A, Hu X, Zheng L, et al. Isolation and characterization of Citrobacter spp. from the intestine of grass carp Ctenopharyngodon idellus. Aquaculture. 2011;313(1-4):156-60.
- Delétoile A, Decre D, Courant S, et al. Phylogeny and identification of Pantoeaspecies and typing of Pantoea agglomerans strains by multilocus gene sequencing. J Clin Microbiol. 2009;2:300-10.
- Warren JR, Farmer JJ, Dewhirst FE, et al. Outbreak of nosocomial infections due to extended-spectrum Ã-lactamase-producing strains of enteric group 137, a new member of the family Enterobacteriaceaeclosely related to Citrobacter farmeri and Citrobacter amalonaticus. J Clin Microbiol. 2000;38(11):3946-52.
- KolÃnská R, Spanelová P, DrevÃnek M, et al. Species identification of strains belonging to genus Citrobacterusing the biochemical method and MALDI-TOF mass spectrometry. Folia Microbiol. 2015;60(1):53-9.
- Clermont D, Motreff L, Passet V, et al. Multilocus sequence analysis of the genus Citrobacter and description of Citrobacter pasteurii sp. Nov. Int J Syst Evol Microbiol. 2015;65(5):1486-90.
- CLSI. Performance standards for antimicrobial susceptibility testing, twenty third informational supplement. CLSI document M100-S23. Wayne, PA: Clinical and laboratory standards institute. 2013;33(1):45-128.
- Nawaz M, Khan AA, Khan S, et al. Isolation and characterization of tetracycline-resistant Citrobacter spp. from catfish. Food Microbiol. 2008;25:85-91.
- Nath G, Maurya P, Gulati AK. ERIC PCR and RAPD based fingerprinting of Salmonella typhi strains isolated over a period of two decades. Infect Genet Evol. 2010;10:530-6.
- Yuan W, Chai TJ, Miao ZM. ERIC-PCR identification of the spread of airborne Escherichia coli in pig houses. Sci total environ. 2010;408:1446-50.
- Macedo NR, Oliveira SR, Lage AP, et al. ERIC-PCR genotyping of Haemophilus parasuis isolates from Brazilian pigs. Vet J. 2011;188:362-4.
- Waturangi DE, Joanito I, Yogi Y, et al. Use of REP- and ERIC-PCR to reveal genetic heterogeneity of Vibrio cholerae from edible ice in Jakarta, Indonesia. Gut Pathog. 2012;4:2.
- Wang W, Gu W, Ding Z, et al. A novel Spiroplasma pathogen causing systemic infection in the crayfish Procambarus clarkii (Crustacea: Decapod), in China. J FEMS Microbiol Lett. 2005;249:131-7.
- Ding Z, Xue H, Xia A, et al. Study on the WSSV prevalence in the cultured crayfish (Procambarus clarkii). J Nanning Agri Univ. 2008;31(4):129-33.
- Chen C, Liu Y, He G, et al. The bacterial pathogen of outbreak disease in Procambarus clarkii. J Huazhong Agri Univ. 2009;28(2):193-197.
- Baumgartner WA, Hawke JP, Bowles K, et al. Primary diagnosis and surveillance of white spot syndrome virus in wild and farmed crawfish (Procambarus clarkii, P. zonangulus) in Louisiana. J Dis Aquat Org. 2009;85:15-22.
- Zhou D, Zhang W, Li J, et al. Isolation and identification of bacterial pathogen from Procambarus clarkii. Chinese Agri Sci Bul. 2011;27(26):102-5.