Research Article - Biomedical Research (2017) Volume 28, Issue 1
Isolation and characterization of novel Tibetan Macaque (Macaca thibetana) microsatellite loci: cross-species amplification and population genetic applications
Na Yang, Liang Zhou, Xiaoshuai Liu and Daiwen Zeng*Institute of Laboratory Animal Science, Sichuan Academy of Medical Sciences & Sichuan Provincial People’s Hospital, Chengdu 610212, Sichuan, PR China
- *Corresponding Author:
- Daiwen Zeng
Institute of Laboratory Animal Science
Sichuan Academy of Medical Sciences & Sichuan Provincial
People’s Hospital
PR China
Accepted on June 02, 2016
Abstract
Aims: To investigate novel microsatellite loci of Tibetan Macaque (Macaca thibetana) and Simple Sequence Repeat (SSR) loci polymorphism in parent population of 30 pairs human and Tibetan Macaques.
Methods: Fluorescence labeling primer PCR and Capillary Electrophoresis were used to detect the Simple Sequence Repeat (SSR) loci polymorphism in parent population of 30 pairs human and Tibetan Macaques.
Result: Results showed that total 12 polymorphism loci were screened out, of which, the Polymorphic information content (PIC) and Expected heterozygosity (HExp) of M. thibetana parent population were 0.718 and 0.761, respectively. The exclude cumulative individual rate was 0.99999 (CNE-I). The average probability that the set of loci was not an unrelated candidate parent from parentage of an arbitrary offspring when the genotype of the other parent was unknown (CNE-1P) and the average probability that the set of loci will not exclude an unrelated candidate parent from parentage of an arbitrary offspring when the genotype of the other parent is known (CNE-2P) were 0.9968 and 0.9999; the genetic diversity of filial generation was slightly lower than that of parents.
Conclusion: The 12 polymorphism loci in M. thibetana were useful for the specific individual identification, effective paternity test and accurate genetic testing.
Keywords
M. thibetana, Genetic diversity, Loci polymorphisms.
Introduction
Tibetan macaque (Macaca Thibetana) is an endemic primate in China. Its geographic distribution range had extended from Western Sichuan through Guizhou, Northeastern Yunnan, Hunan, Northern Guangxi to Anhui, Zhejiang, Jiangxi, Fujian and Northern Guangdong [1]. The average body length of up to 94.70 cm and the heaviest weight can be 30 kg in adult monkey [1]. It is a good new primate for organ transplant since the size and organ shape are similar to humans and pigs in anatomy. M. thibetana was used for the heterotopic auxiliary liver transplantation in three cases of miniature pigs, of which the transplanted liver in vivo function of M. thibetana could be survival for as long as two weeks which had created the current pig-nonhuman primate liver xenotransplantation international longest survival time record [2]. In 2003, artificial breeding was performed to characterize the effects of circadian rhythm, feeding time, age, general anesthesia, and ocular hypotensive compounds on intraocular pressure (IOP) of the M. thibetana [3]. Macaques are the most widely distributed nonhuman primates and used as animal models in biomedical research. The availability of full-genome sequences from them would be essential to both biomedical and primate evolutionary studies. Previous studies have reported whole-genome sequences from rhesus macaque (Macaca mulatta) and cynomolgus macaque (M. fascicularis, CE), both of which belong to the fascicularis group [4]. The particular animal genetic background determines the specificity of their uses; therefore, maintaining their genetic characteristics is the key to ensuring stable and reliable results [5]. Closed feeding practices were used in primates to maintain the genetic diversity of animal populations and avoid inbreeding, which could further improve the long-term viability and animal reproduction rate.
SSR (Simple Sequence Repeats) marker is a kind of specific primers PCR-based molecular marker technology, which is developed in recent years, and it is also known as microsatellite DNA, which is a class of several nucleotides repeated tandem units of up to dozens of nucleotides repeats. The sequences of each SSR sides are generally relatively conservative single-copy sequences [6]. These sequences have been detected in the genomes of numerous organisms and are distributed throughout the entire genome in both coding and non-coding regions [7,8]. Given their many desirable attributes, including wide genomic distribution, co-dominant inheritance, their multi-allelic nature, and a high level of polymorphisms, SSRs are highly favoured molecular markers [9,10]. It has been successfully applied in the field of population genetics non-human primates structural analysis [11-13], paternity [14] and artificial breeding population genetics [5,15]. However, there were few researches about SSR loci in Tibetan macaque reported. In 2011, Jia et al. isolated twenty one microsatellite loci from AC-enriched library of Tibetan macaque and polyacrylamide gel electrophoresis (PAGE) combined with silver stain was used to evaluate the polymorphism of these loci [16]. However, the reliability and efficiency of the silver staining detection is much lower than the fluorescent labeling methods [17]. In the study by Li et al. microsatellite loci were isolated from AAAG-enriched and GATA-enriched libraries of the Tibetan macaque, and the average Polymorphic information content (PIC) and Expected heterozygosity (HExp) were 0.624 and 0.675, respectively [18].
In this study, to detect the genetic characteristics of M. thibetana fastly, simply and sensitively, fluorescence labelling primer PCR and Capillary Electrophoresis were used to detect the SSR loci polymorphism in parent population of 30 pairs human and Tibetan Macaques. Furthermore the applicability of theses loci was evaluated through analysing the genetic characteristics of filial generation. This study would lay a good foundation in the specific individual identification, effective paternity test and accurate genetic testing, and artificial breeding of M. thibetana.
Materials and Methods
Animals
In this research, 81 M. thibetana were raised by institute of laboratory animals of Sichuan academy of medical sciences & Sichuan provincial people’s hospital. Among of them, forty Tibetan monsieur beaucaires were as parents that were captured in mabian county, Sichuan province in 2003 (the relationship is not clear). Others were F1 generations. All animals feeding and management were based on the Chinese animal regulations. The animal experiments were approved by animal ethics committee of institute of laboratory animals of Sichuan academy of medical sciences & Sichuan provincial people’s hospital.
Loci screening and genotyping
Polymerase chain reaction and agarose gel electrophoresis were used to screening the sites. Based on the previous report about human [19], we chosen fifteen sites and corresponding primers. Total DNA was extracted using the Whole Blood Genomic DNA Extraction Kit (TaKaRa, Japan). The PCR reaction was carried out in a total volume of 50 μl containing 50 ng template DNA, 0.25 μM forward and reverse primers, 1×PrimeSTAR Buffer (Mg2+ Plus) Buffer, 1.25U PrimeSTAR HS DNA Polymerase (2.5 U/μl) (PrimeSTAR® HS DNA PolymeraseTaKaRa), 200 μM dNTP. The PCR conditions were as follows: 98°C for 10 seconds, 56-61.3°C for 15 seconds and 72°C for 60 seconds. Based on agarose gel electrophoresis, the corresponding forward primers were fluorescent tags to amplify the whole genome DNA.
According to the PCR products and fluorescence signal strength, the products will be diluted 5 to 50 times, and added 1 μl products to Hi-Di TM methanamide and GeneScanTM500 LIZTM mix. Mixture was turned to the 96-well plates and kept 95°C for 5 minutes, and then put it in ice for 3 minutes. 96- well was put in 3100 genetic analyzer (Applied Biosystems) for capillary electrophoresis. ABI Prism Genescan and Genotyper software were used to screening. Homozygous samples of PCR products of SSR were sequenced (Invitrogen, USA).
Statistical analysis
HObs, HExp, PIC, HW, F (Null), NE-1P, NE-2P, NE-PP, NE-I, CNE-1P CNE-2P, CNE-PP, CNE-I were analyzed using Cervus 3.0.7 software.
Results
Twelve microsatellite loci in exhibited polymorphism M. thibetana
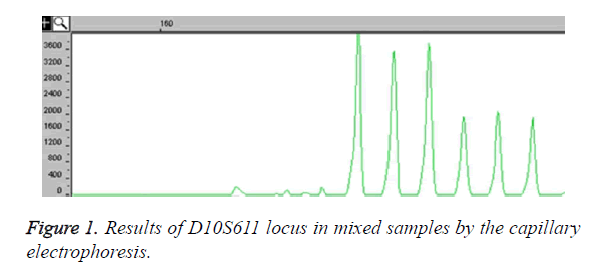
Among the 30 microsatellite loci, a total 24 loci could amplify specific products. In these 24 loci, there are 12 loci having six or more alleles in M. thibetana. Figure 1 showed that the locus D10S611 had polymorphism in parent mixed samples. There have been six alleles in 40 M. thibetana parent samples, and the fragment products sizes were 182 (base pair) bp, 186 bp, 190 bp, 194 bp, 198 bp and 202 bp as it shown in the peak of Figure 1. By sequencing, five loci of the core sequences were two bases repeated, including three loci of single type and two loci of complex type (Table 1). Besides, the results showed that seven loci of the core sequences were four bases repeated, including four loci of single type and three loci of complex type (Table 1).
| Loci | Core Sequence | Loci | Core Sequence | Loci | Core Sequence |
|---|---|---|---|---|---|
| MML3S4 | (TG)15 | MML17S2 | (CA)18 | D09S921 | (GATA)8 |
| MML3S7 | (CA)11 | D03S1768 | (GATA)6(GACA)9 | D10S611 | (TGTC)5(TATC)14 |
| MML3S9 | (TC)19(CA)19 | D05S1457 | (GATA)9 | D13S765 | (GATA)18 |
| MML11S2 | (GT)8(GA)12 | D06S501 | (TATC)10 | D15S823 | (TATG)11(TATC)11 |
Table 1. Core sequence in microsatellite loci.
The screened loci were suitable for the genetics control of Tibetan macaque
A total 93 alleles in 12 loci were screened out in 40 M. thibetana parent samples, and the average HExp and PIC were all above 0.7 (Table 2). When mother-child relationship was unclear, the father identified exclusion probability was 0.99675, while when the mother-child relationship was clear, the exclusion probability was 0.99994 and the individual identification of exclusion probability was 0.99999 (Table 3). Table 2 showed that the locus D15S823 was significantly deviated from Hardy-Weinberg equilibrium because of the overpopulation of homozygote and the null allele possibility was relative high.
| Loci | K | HObs | HExp | PIC | HW | F (Null) |
|---|---|---|---|---|---|---|
| MML3S9 | 14 | 0.975 | 0.867 | 0.841 | ND | -0.0682 |
| D10S611 | 8 | 0.600 | 0.846 | 0.814 | ND | 0.1636 |
| MML11S2 | 9 | 0.700 | 0.829 | 0.797 | ND | 0.0817 |
| D05S1457 | 7 | 0.725 | 0.795 | 0.753 | ND | 0.0390 |
| D15S823 | 8 | 0.359 | 0.781 | 0.740 | * | 0.3640 |
| MML3S7 | 6 | 0.675 | 0.775 | 0.731 | NS | 0.0619 |
| MML3S4 | 10 | 0.564 | 0.735 | 0.702 | NS | 0.1370 |
| D09S921 | 6 | 0.700 | 0.746 | 0.695 | NS | 0.0298 |
| D06S501 | 6 | 0.700 | 0.731 | 0.677 | NS | 0.0190 |
| D13S765 | 6 | 0.600 | 0.712 | 0.651 | NS | 0.0829 |
| D03S1768 | 7 | 0.700 | 0.690 | 0.640 | NS | -0.0114 |
| MML17S2 | 6 | 0.675 | 0.618 | 0.578 | NS | -0.0635 |
| Mean | 7.750 | 0.664 | 0.761 | 0.718 | 0.0697 | |
| SD | 2.379 | 0.141 | 0.070 | 0.077 | 0.1163 |
Table 2. The loci polymorphisms in M. thibetana parent samples (No=40).
| Loci | NE-1P | CNE-1P | NE-2P | CNE-2P | NE-PP | CNE-PP | NE-I | CNE-I |
| MML3S9 | 0.443 | 3.25E-03 | 0.282 | 5.57E-05 | 0.115 | 6.00E-08 | 0.035 | 2.72E-13 |
| D10S611 | 0.503 | 0.331 | 0.159 | 0.049 | ||||
| MML11S2 | 0.526 | 0.351 | 0.170 | 0.055 | ||||
| D05S1457 | 0.599 | 0.420 | 0.238 | 0.079 | ||||
| D15S823 | 0.609 | 0.429 | 0.239 | 0.083 | ||||
| MML3S7 | 0.627 | 0.447 | 0.261 | 0.089 | ||||
| MML3S4 | 0.651 | 0.463 | 0.255 | 0.099 | ||||
| D09S921 | 0.670 | 0.493 | 0.308 | 0.111 | ||||
| D06S501 | 0.687 | 0.512 | 0.327 | 0.122 | ||||
| D13S765 | 0.715 | 0.546 | 0.366 | 0.140 | ||||
| D03S1768 | 0.724 | 0.548 | 0.356 | 0.143 | ||||
| MML17S2 | 0.783 | 0.603 | 0.405 | 0.184 |
Table 3. Individual identification and non-relatives exclusion possibility (No=40).
Genetic diversity between parent and offspring
In order to evaluate the breeding strategy in this research, the 12 loci we screened out were used to detect the genetic diversity in offspring samples. Results showed that comparing with parent samples; there were allele drop-out 6 loci (MML3S9, MML11S2, D05S1457, D15S823, MML3S4 and MML17S2). A total 82 alleles were identified in 41 offspring samples and the HExp and PIC were lower the parents (Table 4).
Discussion
A total of 12 high polymorphic sites in M. thibetana were screened from 30 SSR sites from human and rhesus monkeys. The polymorphic information contents (PIC) were 0.718 (Table 2). The big proportions of heterozygous in these sites indicated that more genetic information is provided. Expected heterozygosity (HExp) is commonly used to measure group of genetic diversity, the higher its value, the lower genetic uniformity of population [19]. The HExp of twelve sites were 0.76 in M. thibetana indicated that the parent population we set up has abundant genetic diversities. What is more, the abundant genetic diversities are also shown in the artificial breeding rhesus monkeys. This suggests that the sites we screened are stable and high polymorphic. However, we also noticed that D15S823 was significantly deviated from Hardy- Weinberg equilibrium. In order to exclude the existence of invalid alleles, we redesigned of primers, results showed that the number of alleles is not changed. The results of Hardy Weinberg equilibrium test may not be reliable since the non-random mating was unknown in the M. thibetana who lived in the field.
Microsatellite markers are powerful tools for paternity test [20], which including NE-1P and NE-2P, and the success rate of NE-2P was higher than NE-1P. In our study the CNE-2P and CNE-1P was 99.99% and 99.68%, which indicated an efficiency paternity test in the screening loci of M. thibetana. Average non-exclusion probability for identity of two unrelated individuals (NE-I) and the average probability that the set of loci will fail to differentiate between two randomly-selected individuals (CNE-I) had the same genotype probability at a particular locus or all loci. The accuracy of site-specific genetic identification of individuals is higher with low CNE-I values. In our study, the CNE-I value was 2.72 × 10-13, which meant that it can identify any one of 3.6×1012 M. thibetana. This number was far more exceeds the current total captivity. Since there was inbreeding in artificial feeding, therefore, the successful rate of paternity test and in individual identification was lower than predicted value. The loci had great value since the number of M. thibetana is very small.
The six alleles in 40 M. thibetana parent samples indicated that the inbreeding was not efficiently avoid, which could be explained by the fact that most of the parents samples were captured in the same place and they belong to one family. It’s of great importance to use microsatellite markers to establish clear and complete pedigree in the breeding process of M. thibetana. In this study, homozygous PCR product of the 12 microsatellite loci were sequenced and we confirmed that 2 and 4 bp repeat were found in core sequence, which was consistent with the other study [19,21]. However, the rhesus microsatellite repeats unit was of unitary type in the literature and there was also complex type in M. thibetana. Among the seven unitary type loci, there were five repeat units which were the same with rhesus. Two nucleotide substitutions exist, but these alleles were not exactly the same as the length of rhesus. This may be due to polymorphism of microsatellite loci in two aspects: the number of nucleotide repeats polymorphism and repeats nucleotide substitution polymorphisms. The nucleotide substitution resulted in the number difference of repeating units and further led to the change in allele length [22]. The length range of complex type loci in M. thibetana was less than rehesus and study showed that complex sequence is likely to be a factor limiting the microsatellite variation [23], which was supported by our study. Although the Tibetan macaque and rhesus species belong to close relatives, however, there were many differences in the microsatellite sequence and numbers in repeating units, which reflecting the complexity of microsatellite evolution.
The sample size was positively correlated with the number of alleles in the detected microsatellite loci [24]. The theoretical minimum sample volume was proposed according to population genetics researchers and probability theory in order to ensure that all the alleles were detected [25]. In our study, MML3S9 was the loci which had the most number of 14 alleles. According to the formula, at least 21 samples were required to ensure that 95% of the alleles were detected, we had 40 samples which were far more beyond the theoretical sample volume and it could completely reflect the loci polymorphism. In conclusion, the 12 polymorphism of microsatellite loci were screened in this work could be used in individual identification, paternity test and genetic tests in M. thibetana. These are reliable and efficient molecular markers that can be applied in the genetics management of M. thibetana.
References
- Jiang XL, Wang YX, Wang QS. Taxonomy and distribution of Tibetan macaque. Zoo Res 1996; 17: 361-369.
- Ji HC, Li X, Yue S, Li J, Chen H, Zhang Z, Ma B, Wang J, Pu M, Zhou L, Feng C, Wang D, Duan J, Pan D, Tao K, Dou K. Pig BMSCs Transfected with Human TFPI Combat Species Incompatibility and Regulate the Human TF Pathway in Vitro and in a Rodent Model . Cell Physiol Biochem 2015.
- Liu G, Zeng T, Yu W, Yan N, Wang H, Cai SP, Pang IH, Liu X. Characterization of intraocular pressure responses of the Tibetan monkey (Macaca thibetana). Mol Vis 2011; 17: 1405-1413.
- Fan Z, Zhao G, Li P, Osada N, Xing J, Yi Y, Du L, Silva P, Wang H, Sakate R, Zhang X, Xu H, Yue B, Li J. Whole-genome sequencing of tibetan macaque (Macaca Thibetana) provides new insight into the macaque evolutionary history. Mol Biol Evol 2014; 31: 1475-1489.
- Kanthaswamy S, Kou A, Satkoski J, Penedo MC, Ward T, Ng J, Gill L, Lerche NW, Erickson BJ, Smith DG. Genetic characterization of specific pathogen-free rhesus macaque (Macaca mulatta) populations at the California National Primate Research Center (CNPRC). Am J Primatol 2010; 72: 587-599.
- Obaid R, Abu-Qaoud H, Arafeh R. Molecular characterization of three common olive (L.) cultivars in Palestine, using simple sequence repeat (SSR) markers. Biotechnol Biotechnol Equip 2014; 28: 813-817.
- Li YC, Korol AB, Fahima T, Beiles A, Nevo E. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol 2002; 11: 2453-2465.
- Wu YQ, Huang Y. An SSR genetic map of Sorghum bicolor (L.) Moench and its comparison to a published genetic map. Genome 2007; 50: 84-89.
- Xiao J, Zhao J, Liu M, Liu P, Dai L, Zhao Z. Genome-Wide Characterization of Simple Sequence Repeat (SSR) Loci in Chinese Jujube and Jujube SSR Primer Transferability. PLoS One 2015; 10: e0127812.
- Hughes CR, Queller DC. Detection of highly polymorphic microsatellite loci in a species with little allozyme polymorphism. Mol Ecol 1993; 2: 131-137.
- Oliveira CG, Gaiotto FA, Costa MA, Martinez RA. Molecular genetic analysis of the yellow-breasted capuchin monkey: recommendations for ex situ conservation. Genet Mol Res 2011; 10: 1471-1478.
- Hagell S, Whipple AV, Chambers CL. Population genetic patterns among social groups of the endangered Central American spider monkey (Ateles geoffroyi) in a human-dominated landscape. Ecol Evol 2013; 3: 1388-1399.
- Baden AL, Holmes SM, Johnson SE, Engberg SE, Louis EE Jr, Bradley BJ. Species-level view of population structure and gene flow for a critically endangered primate (Varecia variegata). Ecol Evol 2014; 4: 2675-2692.
- Newman TK, Fairbanks LA, Pollack D, Rogers J. Effectiveness of human microsatellite loci for assessing paternity in a captive colony of vervets (Chlorocebus aethiops sabaeus). Am J Primatol 2002; 56: 237-243.
- Ogata M, Seino S. Genetic analysis of captive proboscis monkeys. Zoo Biol 2015; 34: 76-79.
- Jia XD, Yang BD, Yue BS, Yin HL, Wang HX, Zhang XY. Isolation and characterization of twenty-one polymorphic microsatellite loci in the Tibetan macaque (Macaca thibetana). Genetika 2011; 47: 996-999.
- Hao CY, Wang LF, Jia JZ, Dong YC, Zhang XY. Comparison of Fluorescence and Silver-staining Detection Systems of Microsatellite Markers. Acta Agronomica Sinica 2005; 31: 144-149.
- Li P, Yang CZ, Zhang XY. Development of eighteen tetranucleotide microsatellite markers in Tibetan Macaque (Macaca thibetana) and genetic diversity analysis of captive population. Biochemical Systematics and Ecology 2014; 57: 293-296.
- Kanthaswamy S, von Dollen A, Kurushima JD, Alminas O, Rogers J, Ferguson B, Lerche NW, Allen PC, Smith DG. Microsatellite markers for standardized genetic management of captive colonies of rhesus macaques (Macaca mulatta). Am J Primatol 2006; 68: 73-95.
- Smith DG, Kanthaswamy S, Viray J, Cody L. Additional highly polymorphic microsatellite (STR) loci for estimating kinship in rhesus macaques (Macaca mulatta). Am J Primatol 2000; 50: 1-7.
- Raveendran M, Harris RA, Milosavljevic A, Johnson Z, Shelledy W, Cameron J, Rogers J. Designing new microsatellite markers for linkage and population genetic analyses in rhesus macaques and other nonhuman primates. Genomics 2006; 88: 706-710.
- Garza JC, Slatkin M, Freimer NB. Microsatellite allele frequencies in humans and chimpanzees, with implications for constraints on allele size. Mol Biol Evol 1995; 12: 594-603.
- Morin PA, Mahboubi P, Wedel S, Rogers J. Rapid screening and comparison of human microsatellite markers in baboons: allele size is conserved, but allele number is not. Genomics 1998; 53: 12-20.
- Maudetr C, Miller C, Bassano B, Breitenmoser-Würsten C, Gauthier D, Obexer-Ruff G, Michallet J, Taberlet P, Luikart G. Microsatellite DNA and recent statistical methods in wildlife conservation management: applications in Alpine ibex [Capra ibex(ibex)]. Mol Ecol 2002; 11: 421-436.
- Von Segesser F, Menard N, Gaci B, Martin RD. Genetic differentiation within and between isolated Algerian subpopulations of Barbary macaques (Macaca sylvanus): evidence from microsatellites. Mol Ecol 1999; 8: 433-442.