Research Article - Journal of Biochemistry and Biotechnology (2022) Volume 5, Issue 6
Isolation and Characterization of non-tuberculous mycobacteria from water in Ahmadu Bello University Teaching Hospital, Zaria.
F. B. Tanimu1*, A. Lawal3, A. Balarabe2, H.G Anchau1 and J.M Yelwa1
1Scientific and Industrial Research department, National Research Institute for Chemical Technology, Zaria, Nigeria
2National Agricultural Extension and Research Liaison services, Ahmadu Bello University, Zaria
3Department of Vet., Public Health and Preventive Medicine, Bayero University Kano, Janbulo Second Gate Rd, Nigeria
- Corresponding Author:
- Tanimu FB
Scientific and Industrial Research Department
National Research Institute for Chemical Technology
Zaria, Nigeria
E-mail: ftbalarabe94@gmail.com
Received: 12-Aug-2022, Manuscript No. AABB-22-71771; Editor assigned: 15-Aug-2022, PreQC No. AABB -22-71771(PQ); Reviewed: 29-Aug-2022, QC No AABB-22-71771; Revised: 02-Sep-2022, Manuscript No. AABB-22-71771(R); Published: 12-Sep-2022, DOI:10.35841/aabb-5.5.122
Citation: Tanimu FB, Lawal A, Balarab A, et al. Isolation and characterization of non-tuberculous mycobacteria from water in Ahmadu Bello University Teaching Hospital, Zaria. J Biochem Biotech 2022;5(5):122
Abstract
Non tuberculous mycobacteria (NTM) found frequently in environmental samples cause important opportunistic infections in immunocompromised patients. The aim of the study was to isolate and characterize non tuberculous mycobacteria in water samples from Ahmadu Bello University teaching hospital Zaria. A total of 32 water samples were collected from 13 different locations. The samples were processed by homogenization and decontamination, culture on Lowenstein Jensen medium and acid-fast staining was performed. Acid fast bacilli isolated from culture medium were identified using conventional biochemical tests which are of limited value in differeciation of NTM species. NTM were identified from 3(9.375%) of the 32 examined samples. Two of the three NTM positive samples were stored water samples collected from HIV Clinic and mosque. It was concluded that NTM contamination of water and environment in hospital was a potential risk factor in terms of nosocomial infections. Thus, surveillance cultures of water in the hospital are necessary to fix the source of NTM, to identify the types and strains and to establish control measures such as sterilization, disinfection, maintenance and modernization of water systems.
Keywords
MacConkey, Inoculation, Organism, Microscopic, Environmental.
Introduction
Non-tuberculous mycobacteria (NTM) are widely present in the environment and are commonly isolated from environmental sources including water [1]. Several species of environmental mycobacteria have been known to be important human pathogens, and reports suggest that there is an increasing trend in the incidence of NTM disease [2,3], and in particular, in immunocompromised patients [4,5]. Exposure to NTMs occurs primarily through human interactions with water [3]. In particular, NTM species can multiply in the numerous water sources, including waste water, surface water, ground water, and tap water [6]. Mycobacteria are able to multiply within low nutrient environment particular!}" Water pipe systems and can survive in hospital hot water systems, and resist chlorination. Moreover, biofilm formation, amoeba-associated lifestyle, and resistance to chlorine have been recognized as important factors that contribute to the survival, colonization and persistence of NTM in water distribution systems [7,8]. The incidence of NTM has been reported different from one setting to another, ranging from 40 [9] to 58% [10]. Some aquatic systems such as hospital water lines are more suitable for colonization with mycobacteria and biofilm formation [1].
Materials and Methods
Sample size
A total of 32 water samples were collected from Ahmadu Bello University teaching hospital Zaria.
Sample collection
Samples were collected in a clean sterile container using hand gloves and then samples were transported to the laboratory for experiment.
Materials, media and reagents
Glass-wares such as test tubes and micropipettes, universal container, centrifuge. 4%NaOH, Ziehl-Neelsen staining reagents and biochemical test reagent were used.
Preparation of culture media
About 37.7g of Lowenstein Jensen medium was dissolved in 600 ml of cold Purified Water containing 12 ml of glycerol. It was heated to boiling with constant agitation and autoclave at 121°C for I5 minute. A uniform suspension of fresh eggs (1000 ml) was prepared under aseptic conditions; whipping air into suspension will be avoided during the collection and mixing. The 1000 ml of egg suspension was aseptically mix with the 600 ml of the sterile Lowenstein Jensen medium Base cooled between 45ºand 60°C avoiding air bubbles. The finished medium was dispensed into sterile screw-cap test tubes and the test tubes were placed in a slanted position and heated for 85°C for 45minute (Sagar, 2015) (Figure 1).
Inoculation of sample
Water samples, 50ml each were collected from water sources. Water sample (total volume) was centrifuged at 3,000 xg for 15 min at 4°C and the supernatant was discarded. Pellets from the water samples were suspended separately in 20 ml of 4% NaOH and incubated at room temperature for 30 min. After incubation, about 20 ml of phosphate buffer was added and incubated for 5 min. the suspensions were centrifuged at 3,000 xg for 15 min at 4°C, and the supernatants was decanted. The pellet was finally re-suspended in 2 ml of phosphate buffer. One ml of the suspension was inoculated into Lowenstein-Jensen media and incubated for 1-2 weeks. After incubation, the colonial morphology was observed for smooth or intermediate, humid, yellow, orange or creamcolored colonies, it was stained using Ziehl Neelsen technique and sub-cultured on Lowenstein-Jensen medium to obtain pure culture (Figure 2) [11].
Ziehl Neelsen staining procedure
The wire loop was flamed to red hot. allowed to cool then a loop full of sample was picked and placed on a clean grease free slide and a smear made, air dried and heat fixed by passing smear over flame for 3 times. The glass slide was placed on a staining rack and smear covered with 1% carbon fuschin solution and heated to steam, it was allowed to stay for 5 mins and rinsed with water, then 3% acid alcohol-solution was applied on the same slide for another 3mins and finally rinsed with water under a slow running tap, then the smear was flooded with methylene blue and allowed to stay for 1 min after which the slide was rinsed in slow running tap water. The slide was kept in slant position to drain and air dry, slides were then carefully observed microscopically by placing a drop of oil immersion on the smear and viewed using the oil immersion objective (x100) [11] (Figure 3).
Catalase test
Several loopful of growth was suspended into a screw capped tube containing 0.5 ml of buffer solution (PH 7.0), The suspension was incubated in a 68°C water bath for 20 minutes and then cooled to room temperature 0.5 ml of the freshly prepared tween-peroxide mixture was added and recapped loosely. The formation of bubbles appearing on the surface of the hand was observed [11].
Nitrate reduction test
One loopful of organisms from culture was suspended into 2 ml of the buffered solution sodium nitrate. The culture was allowed to incubate in a 37°C water bath for 2hour. After which time the suspension was acidified with 1 drop of HCL solution. Then subsequent addition of 2 drops sulfanilamide solution and 2 drops naphtyldiethyl dihydrochloride results in the immediate formation of fuchsia pink color [11] (Figures 4-6).
Urease test
A loopful of the organism was used to inoculate the entire surface of the urea slant. The cap was tightened and incubated at 37°C for 24-48 hours and was observed for pink color change [11].
Growth on MacConkey
The organism was inoculated directly on the surface of MacConkey agar plate and streaked for isolation. It was incubated at 35-37°C for 48-12 hours and was examined for growth [11].
Result
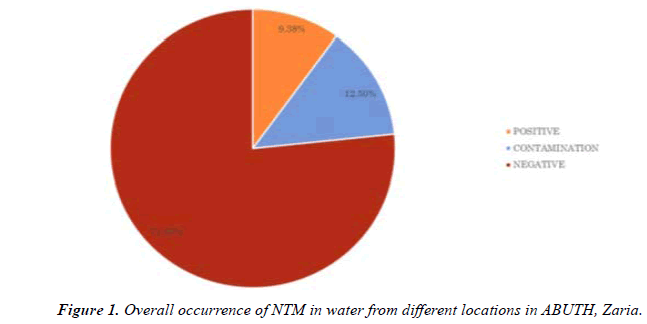
NTM were isolated from 3 of the 32 samples analyzed (9.375%) (Table 1). And from two of the four sources examined (Table 2). NTM were not isolated from tap water and tank water (Table 3).
| Isolate code | Colonial morphology on LJ medium | Growth on MacConkey agar | AFB staining | Biochemical test |
Inference | ||
|---|---|---|---|---|---|---|---|
| CAT | U | NR | NTM | ||||
| A | SMOOTH YELLOW-ORANGE | + | + | NEGATIVE | |||
| B | NO GROWTH | _ | _ | + | + | + | |
| _ | _ | _ | |||||
Table 1. Cultural, Microscopic and Biochemical Characteristics of the isolates.
| S/N | SAMPLING AREA | NUMBER OF SAMPLE | NUMBER OF NTM POSITIVE SAMPLE (%) |
|---|---|---|---|
| 1 | NASARA | 4 | 1(3.13%) |
| 2 | MOSQUE | 4 | 1(3.13%) |
| 3 | KITCHEN | 4 | 0(0.00%) |
| 4 | EMERGENCY | 2 | 0(0.00%) |
| 5 | MALE HOSTEL | 2 | 0(0.00%) |
| 6 | FEMALE HOSTEL | 2 | 1(3.13%) |
| 7 | LAUNDARY | 2 | 0(0.00%) |
| 8 | TRANSIENT CAMP | 2 | 0(0.00%) |
| 9 | MALE SURGICAL | 2 | 0(0.00%) |
| 10 | FEMALE SURGICAL | 2 | 0(0.00%) |
| 11 | MALE ORTHOPEDICS | 2 | 0(0.00%) |
| 12 | FEMALE ORTHOPEDICS | 2 | 0(0.00%) |
| 13 | HAEMATOLOGY | 2 | 0(0.00%) |
Table 2. Occurrence of NTM based on sampling area.
| S/N | SPECIFIC SAMPLING LOCATION | NUMBER OFSAMPLE COLLECTED | NUMBER OF NTM POSITIVE SAMPLE (%) |
|---|---|---|---|
| 1 | TANK WATER | 8 | 0(0.00%) |
| 2 | BOREHOLE | 8 | 1(3.13%) |
| 3 | TAP WATER | 8 | 0(0.00%) |
| 4 | STORED WATER | 8 | 2(6.25%) |
Table 3. Occurrence of NTM based on specific sampling locations.
Discussion
In this study, phenotypic methods were applied for the identification of NTM species in hospital water samples. Rapidly growing NTM were isolated from 3 (9.375%) samples and identified using the scheme provided by Wellman, (2012). This agrees with previous studies which showed that NTM are widely present in the environment and are commonly isolated from soil and water [12].
The overall occurrence of NTM was found to be 3(9.38%). This is lower than the percentage occurrence (16%) reported from Mexico by Fernandez et al., (2012). This may be due to the lower number of examined samples and method of detection. Though the phenotypic method of identification of NTM is of limited value in differeciation of NTM species, [13] it is used for preliminary identification of their presence in environmental samples. Sequence based method offer much more resolution in identification of NTM compared to other assays where the isolates will be identified correctly to the species level.
Report from Iran has demonstrated M. fortuitum as dominant species both in water and clinical samples [13,14] in contrast, in several study M gordonae, M kansasti and M chelonae were shown to be the most prevalent mycobacterial species in water supplies of Hospital as reported by [9,15-17] NTM occurred more in stored water (6.25%) and less in borehole (3.13%) This may be due to use of contaminated containers for storage, hand-water contact and sanitary condition of the environment [18-20].
Conclusion
The findings of the present study suggest that water is an important environmental source harboring NTM. The overall occurrence of NTM was found to be 3(9.38%).
References
- Decker BK, Palmore TN. The role of water in healthcare-associated infections. Curr opin Infect Dis. 2013;26(4):345.
- Moore JE, Kruijshaar ME, Ormerod LP, et al. Increasing reports of non-tuberculous mycobacteria in England, Wales and Northern Ireland, 1995-2006. BMC public health. 2010;10(1):1-6.
- Donohue MJ, Mistry JH, Donohue JM, et al. Increased frequency of nontuberculous mycobacteria detection at potable water taps within the United States. Environ Sci Technol. 2015;49(10):6127-33.
- Parashar D, Chauhan DS, Sharma VD, et al. Optimization of procedures for isolation of mycobacteria from soil and water samples obtained in northern India. Appl Environ Microbiol. 2004;70(6):3751-3.
- Narang R, Narang P, Mendiratta DK. Isolation and identification of nontuberculous mycobacteria from water and soil in central India. Indian J Medl Microbiol. 2009;27(3):247-50.
- Edirisinghe EA, Dissanayake DR, Abayasekera CL, et al. Occurrence of nontuberculous mycobacteria in aquatic sources of Sri Lanka. Int J Mycobacteriol. 2014;3(4):242-6.
- Vaerewijck MJ, Huys G, Palomino JC, Swings J, Portaels F. Mycobacteria in drinking water distribution systems: ecology and significance for human health. FEMS Microbiology Rev. 2005;29(5):911-34.
- Falkinham JO. Environmental sources of nontuberculous mycobacteria. Clin Chest Med. 2015;36(1):35-41.
- Shin, J.H, Lee, E.J, Lee, H.R, et al. Prevalence of non-tuberculous mycobacteria in a hospital environment. Environmental Microbiology. 2007;65:143-148.
- Crago B, Ferrato C, Drews SJ, et al. Surveillance and molecular characterization of non-tuberculous mycobacteria in a hospital water distribution system over a three-year period. J Hosp Infect. 2014;87(1):59-62.
- Washington JA (2012). Laboratory procedurs in clinical microbiology 2nd edtion springer-verlag.Ne wyork. 3 85-399.
- M, Lamei A, Babazadeh H, Yavari SA. Isolation of rapid growing mycobacteria from soil and water in Iran. Afr J Biotechnol. 2010;9(24):3618-21.
- Moghim S, Sarikhani E, Esfahani BN, et al. Identification of nontuberculous mycobacteria species isolated from water samples using phenotypic and molecular methods and determination of their antibiotic resistance patterns by E-test method, in Isfahan, Iran. Iran J Basic Med Sci. 2012;15(5):1076.
- Shojaei H, Heidarieh P, Hashemi A, et al. Species identification of neglected nontuberculous mycobacteria in a developing country. Japanese J Infectious Diseases. 2011;64(4):265-71.
- Sebakova H, Kozisek F, Mudra R, Kaustova J, Fiedorova M, Hanslikova D, Nachtmannova H, Kubina J, Vraspir P, Sasek J. Incidence of nontuberculous mycobacteria in four hot water systems using various types of disinfection. Can J Microbiol. 2008;54(11):891-8.
- Hussein Z, Landt O, Wirths B, et al. Detection of non-tuberculous mycobacteria in hospital water by culture and molecular methods. Int J Med Microbiol. 2009;299(4):281-90.
- Montanari LB, Sartori FG, Cardoso MJ, et al. Microbiological contamination of a hemodialysis center water distribution system. Rev Inst Med Trop Sao Paulo. 2009;51:37-43.
- Adékambi T, Colson P, Drancourt M. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J Clin Microbiol. 2003;41(12):5699-708.
- Ribón W. Biochemical isolation and identification of mycobacteria. Biochemical Testing. InTech. 2012 Mar 7:21-52.
- Hashemi-Shahraki A, Bostanabad SZ, Heidarieh P, et al. Species spectrum of nontuberculous mycobacteria isolated from suspected tuberculosis patients, identification by multi locus sequence analysis. Infect Genet Evol. 2013;20:312-24.
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref