Review Article - Asian Journal of Biomedical and Pharmaceutical Sciences (2021) Volume 11, Issue 81
In-silico pharmacological studies and RMSD comparison of FDA drugs with bioactive compounds in musa paradisica for ligand promotion.
Rakesh N R*, Gurumurthy H, Pradeep S, Kumar H M, Harshitha, Yashaswini
Department of Biotechnology, BMS College of Engineering, Karnataka, India
- Corresponding Author:
- Dr. Rakesh NR
Department of Biotechnology BMS College of Engineering
Karnataka
India
Tel: 9900696111
E-mail: rakeshnr@gmit.ac.in
Accepted date: 20 September, 2021
Abstract
This study includes Bio-Pharmaceutical research for atomic demonstrating techniques, inside an assortment of medication revelation programs, to examine complex organic and compound frameworks. The coordination of computational and test procedures has been of extraordinary incentive in the distinguishing proof and advancement of novel promising mixes. Comprehensively utilized in a present-day drug plan, sub-atomic docking techniques investigate the ligand compliances received inside the coupling locales of macromolecular targets. We are considering Musa species for our medication revelation measure by utilizing bioinformatics programming like Chemspider, Swiss ADME, Swiss prediction to find out the analogues of a particular compound following their related physicochemical properties including both primary and secondary properties for the structure-based medication plan. Finally leading to evaluate the suitable drug compound present in Musa species for the drug design using Computer-Aided Drug Design (CADD).
Keywords
Chem spider, Swiss ADME, Computer aided drug discovery, Molecular refractivity,Drug- likeness.
Introduction
Pharmaceutical research has successfully adopted the molecular modelling methods for drug discovery process and programs. It is used to study the complexity of chemical and biological compounds. These experimental strategies and computational methods helps for development and identification of novel compounds. In modern drug designing process this computational strategies are broadly used. Molecular docking method helps to understand the stable ligand conformations between one molecules to the other along the active binding sites of target proteins.
Musa is one of the a few genera in the family Musa-ceae. It incorporates bananas and plantains. Plantains allude in India to a coarse banana. Musa has been cultivated for quite 4000 years, the first varieties are increased to 300. Around 70 types of Musa are known, with a wide assortment of application. Musa is a plant species in which we can see both female and male organs in the same individual. This plant has large, broad green leaves, bears flowers and fruits, and grows up to 40 feet in height. These leaves grow through a hollow stem. This gathering of plants are ordinarily developed in all the tropical territories and generally found in nations like India, Burma, Australia, tropical Africa and America.
Musa species are well known for several common Human health biological beneficial activities. One of the conventional source form usually done from ancient medication. The various components of the plant are used for the treatment of many disorders. This technique is citied and restricted to Egypt, Florida, South Brazil, and Southern Japan. For the treatment of respiratory disease and for cough issues its leaves are used. And its roots for the treatment of hemoptysis arrest and used as an Anthelmintic. Its fruits reduce the chance of urinary organ cancer and additionally increase the urinary organ activities within the body. This can be additionally effective in reducing the consequences of free radicals if antioxidants are present in it. It can likewise be utilized as a counteracting for consumes, tuberculosis, fever, asthma, gout, syphilis, snake bite, and warts. These plant components can even be used for the immune system strengthening, to boost muscular activity, lack of appetence, to scale back the mental shock and stomach ulcers, and to own robust bone health.
To be a good drug, the molecule ought to reach the target web site within the body and stay there for a extended time in its bioactive kind, and it ought to be in sufficient concentration. Once it reaches the location it ought to be able to perform the biological events to cure or cut back the matter within the body. In early medication disclosure and improvement technique the appraisal of ADEM are concerned. During this process we’ve to think about several compounds however there physical samples are restricted. So, as an optional computer models of this compounds and constitute are used for additional experiments.
SMILES signify Simplified Molecular Input Line Entry System. It’s illustration of a molecule or a matter within the variety of line notation. SMILES strings are vital for the molecule editors for the conversion of these line notations into 2-D drawings or to 3-D models of the molecules. And molecular graph may be illustration of structural formulas of the chemical compounds in terms of graph theory.
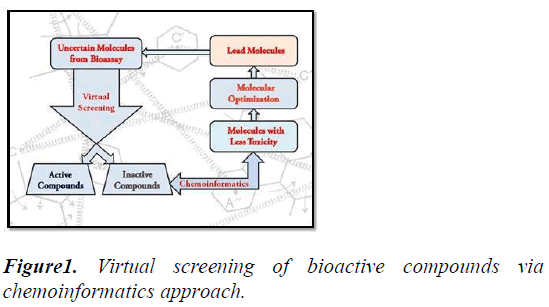
Using these chemical structures the various molecular descriptors has been developed by Chem informaticians. Molecular fingerprint (FP) may be a one in all the favoured example; it consists of sequence bits that facilitate United States of America to spot the absence and presence or changes in chemical structure of the molecules. The FP2 method is one in all the trail based mostly FP, it considers the complete bond present within the molecular structure to get range of bonds. And therefore the physicochemical descriptors facilitate United States of America to get the common properties like mass (MW), and specific atom sorts and their total counts (Figure 1).
Figure 1: Virtual screening of bioactive compounds via chemoinformatics approach.
“Drug-likeness” is employed to understand the probabilities of a molecule to become a lead molecule within the formulation of oral drug with regard to bioavailability. Bioavailability measures the rate at which a drug reaches a web site of action. Drug-likeness is studied to understand the thoughts that compounds have the flexibility to be a oral drug-candidate. And this finished the inspections of structural and physicochemical properties of developed compounds, provided that all the parameters like TPSA, Molar refractivity, LOGP values, LOGS, Solubility category, Bioavailability score, Lead likeness, Lipinski’s rule and Synthetic accessibility are satisfied and the compound can be considered as a lead molecule for the event of the oral drug. This routine is employed to filter the chemical libraries to get rid of the molecules that are incompatible or not within the appropriate pharmacological medicine profile.
In drug discovery process molecular docking and molecular modeling has become one among the main steps. The docking study may be a process during which study of bonds is conducted, ligand formations and bindings between one molecule to a different is studied to get a stable chemical complex. There are different molecular docking studies in drug discovery pipe line, development and applications of this method in drug discovery can’t be neglected. The molecular docking approach is employed to model the interactions between proteins and a molecules in an atomic level, it help us to predict the reaction of molecules once they bind to the target protein sites. With these studies we will conclude the biochemical process administered during the protein and molecule interaction.
The docking includes two essential advances:
• Preparation and Prediction the conformation of Ligand and
• Its position and direction inside these destinations and evaluation of the Interaction affinity.
These two steps are associated with sampling methods and scoring schemes, respectively. During the docking studies knowing the target protein binding site increases the efficiency of the results obtained. Target identification is process where pattern matching for sequences, computational protein annotation, impact of genetic variation on drug discovery and development are studied. And target validation is that the study of pathways and networks, molecular interactions in protein complexes. Basically, the significant point of atomic docking is to offer a right expectation of the ligand-receptor complex structure of compound and protein restricting utilizing calculation techniques. Docking results are acquired with two stages: first by ligand compliance of the testing inside the site of the protein at that point positioning these adaptations through a scoring capacity.
Objectives:
• To identify the compounds present in the Musa species for Analog search.
• Analysis and Prediction of different Pharmological Properties of Bioactive Compounds
• Identification of Similar kind of FDA Approved Drugs and Biological Target Prediction
• Evaluation of Predicted Bioactive compound via Computer Aided Drug Discover (CADD).
Literature Review
A review: pharmacognostic studies and pharmacological actions of Musa paradisiaca
A review article by the authors D.Swathi1, B.Jyothi and C.Sravanthi. During this paper we’ll find that every part (leaves, ripe and unripe fruits, stems) of Musa Paradisiaca contain bioactive compounds which are utilized in Ayurveda for the treatment of snake bite, Insomnia, Hypertension, Diabetes and Asthma. We’ll also obtain several experimental studies of multiple biological activities from this paper [1].
A phytochemical and pharmacological review
This is a review paper written by Mohammad Zafar Imam and Saleha Akter from Bangladesh. The authors reviewed many papers and concluded that the phytochemicals present in Musa paradisiaca L. and Musa sapientum L. are traditionally used to treat diabetes, gout, diarrhoea, cardiac disease, uraemia and hypertension etc,. During this paper we’ll see the reports on phytochemicals identified and isolated from the flowers, seeds, peels and fruit pulp of the plant and also the biological activities of the varied bio-compounds extracted. We’ll also obtain the traditional uses and scientific information, pharmacology and phytochemistry of the compounds present in these two species [2].
Adaptogenic studies of acetone extract of Musa paradisiaca L. fruit peels in albino Wistar rats
Authors of this paper are Sibi Ittiyavirah, D. A. Anurenj from Kerala, India. The acetone extract of plantain unripe fruit peels and acetone extracts of plantain ripe fruit for the evaluation of antistress activity is studied. The high amount of triterpenoids and plant sterols are expected to reduce the chronic depression and in modulating the effect of cholesterol during stress. The high amount of antioxidants present within the peel extract is used to produce the adaptogenic activity within the body. The experiment is conducted for 3 weeks. Both the samples confirmed to possess the antidepressant activity. Hence it’s concluded that the acetone extract of both unripe and ripe fruit peel had shown promising antistress activity [3].
Molecular docking and structure-based drug design strategies.
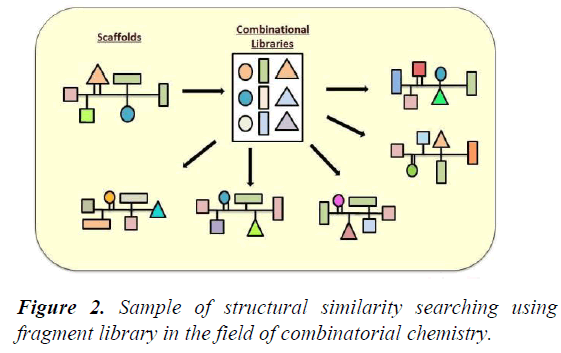
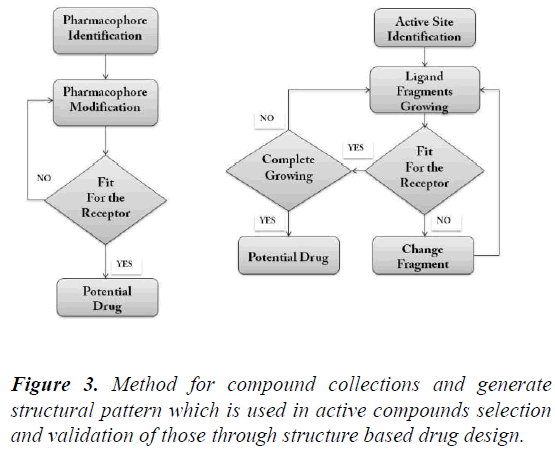
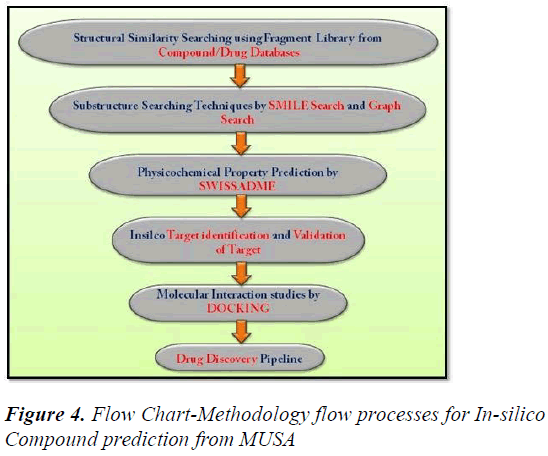
A review paper written by Leonardo G. Ferreira, Ricardo N. dos Santos, Glaucius Oliva and Adriano D. Andricopulo. Paper consists of knowledge on the molecular modeling methods in drug discovery programs. We’ll study the methods used for the event and identification of the novel compounds for drug formulation. Docking methods, target validation, target identification, adaptation of compound and protein target site to urge the stable complex structure. Effectiveness of all the methods and their limitations can also be obtained from this review paper and also information on how to recognize the intermolecular interaction phenomena. The review paper contents include current molecular docking strategies utilized in medicinal chemistry and drug discovery, and also advances in structure based drug designing and ligand based drug designing methods [4] (Figure 2-Figure 5).
Figure 2: Sample of structural similarity searching using fragment library in the field of combinatorial chemistry.
Figure 3: Method for compound collections and generate structural pattern which is used in active compounds selection and validation of those through structure based drug design.
Figure 4: Flow Chart-Methodology flow processes for In-silico Compound prediction from MUSA
Figure 5: Structural similarity searching using fragment library from compound/drug databases.
ChemSpider is a tool used for similarity searching and the analogs are chosen for their further studies (Figure 6) [5].
Figure 6: Substructure searching techniques by smile search and graph search.
In the Figure SwissADME tool is used for finding SMILES of a particular compound by Uploading a 3D structure of the required compound (Figure 7).
Figure 7: Physicochemical property prediction by SWISSADME.
Figure indicating physicochemical properties of a given compound which comprises of both essential and optional properties. Essential properties incorporate SMILES, Molecular formula and weight, Number of Rotatable Bonds, Number of hydrogen Bond acceptors and donors, Topological Polar Surface Area along with Secondary properties like Solubility, Leadlikeness, Lipinski property, Bioavailability score (Table 1).
| Sl. No | Assigned | Compounds |
|---|---|---|
| Molecular-ID | ||
| 1 | 1A1.1 | Sitosterol |
| 2 | 2A1.2 | Daucosterol |
| 3 | 3A1.3 | Fucostanol |
| 4 | 4A1.4 | β-Sitosterol acetate |
| 5 | 5A1.5 | clionasterol |
| 6 | 6A2.1 | (2R,3S,4S)- leucocyanidin |
| 7 | 7A2.2 | Leucocianidol |
| 8 | 8A3.1 | Syringin |
| 9 | 9A4.1 | Quercetin |
| 10 | 10A5.1 | DL-Tryptophan |
| 11 | 11A5.2 | L-5-hydroxy-Tryptophan |
| 12 | 12A5.3 | DL-N-Acetyltryptophan |
| 13 | 13A6.1 | Serotonin |
| 14 | 14A6.2 | Bufotenine |
| 15 | 15A7.1 | Isochroman |
| 16 | 16A7.2 | 1,2,3,3a- Tetrahydropyrrolo[1,2- a]quinoxalin-4(5H)-one |
| 17 | 17A7.3 | 7-Hydroxy-5-methoxy-2- phenyl-chromen-; 4-one |
| 18 | 18A7.4 | 5-(4-Chloro- benzylidene)-3-ethyl-2- thioxo-thiazolidin-4-one |
| 19 | 19A7.5 | 4-oxo-nonenal |
Table 1. List of bioactive compound from Musa and their assigned molecular-id.
Results and Discussion
Table shown below consists of the compounds in the Musa species and the analogs obtained from the combinational libraries with their 3D modeling (Table 2) [5].
| Model Protein-ID | Template PDB-ID | % Similarity | Protein Class/ Significance |
|---|---|---|---|
| P05023 | 4xe5.1.A | 97.45 | Sodium/ potassium- transporting ATPase subunit alpha-1 |
| P06401 | 1sqn.2.A | 100 | progesterone receptor |
| P10275 | 1e3g.1.A | 100 | ANDROGEN RECEPTOR |
| Q7Z2W7 | 6o77.1.A | 82.66 | Transient receptor potential cation channel subfamily M member 8 |
| P35610 | 6bug.1.B | 12 | D-alanyl transfer protein DltB |
| P35611 | 3ocr.1.A | 44 | Class II aldolase/ adducin domain protein |
| P44469 | 6g9p.1.A | 59 | Peptidoglycan D,D- transpeptidase MrdA |
| P11712 | 5x24.1.A | 99 | Cytochrome P450 2C9 |
| Q9RVD6 | 2a4m.1.B | 100 | Tryptophanyl- tRNA synthetase II |
| Q9UGM6 | 5ekd.1.A | 100 | Tryptophan--tRNA ligase, mitochondrial |
| P48039 | 6me2.1.A | 95 | chimera protein of Melatonin receptor type 1A and GlgA glycogen synthase |
| P48040 | 6me8.1.A | 62 | Soluble cytochrome b562,Melatonin receptor type 1B,Rubredoxin |
| P10613 | 5v5z.1.A | 100 | Lanosterol 14- alpha demethylase |
| P22303 | 6o5s.1.A | 100 | Acetylcholinestera se |
| Q9Y5Y9 | 6j8e.1.B | 62 | Sodium channel protein type 2 subunit alpha |
| Q12791 | 6j8e.1.B | 62 | Sodium channel protein type 2 subunit alpha |
Table 2. List of modeled protein-ID with targets template PDB- ID along with percentage (%) of similarity and their class/ significance.
The 3D model, when transferred to the SWISSADME instrument certain physicochemical properties, are viewed as, for example, Sub-atomic Formula close by its Biomolecular weight, Simplified Molecular Input Line Entry System gives a line documentation for encoding sub-atomic structures and explicit examples, Topological Polar Surface Area which giving the surface total over all the polar iotas or particles essentially oxygen and nitrogen including their joined hydrogen ions (TPSA between 20 and 130 Å2 ), Molecular Refractivity is the proportion of absolute polarizability of a mole of a substance and is subject to temperature, file of refraction and weight. Log S (ESOL) for predicting the intrinsic water solubility by using log S scale: insoluble<−10<poorly<−6<moderately<−4<soluble<−2<very<0<highly. Lipophilicity is the partition coefficient between n-octanol and water (log Po/w) which is the classical descriptor.
Lipinski’s rule is a thumb rule to evaluate drug likeness or to determine if the chemical compound having pharmacological activity has chemical or physical properties that would
make it a likely orally active drug in humans.
The parameters to be followed in Lipinski’s rule are MW<=500, MLOGP<=4.15, N or O<=10, NH or OH<= 5.
Drug- Likeness evaluates qualitatively a chance for a molecule to become an oral drug with respect to its bioavailability and helps in the filtering of chemical libraries to eliminate the molecules which are most probably incompatible with an acceptable pharmacokinetic profile. Bioavailability Score is for the rapid appraisal of drug-likeness, the score is 0.11 for anions for which TPSA is >150 Å 2, 0.56 if TPSA is between 75 and 150 Å 2, and 0.85 if TPSA is <75 Å
2. Lead likeness is considered as it helps to obtain the compound which has the desired properties to be a lead molecule in the development of drug, the parameters considered are (250<=MW<=350, XLOGP<=3.5, rotatable bonds<=7). Synthetic Accessibility (SA) Score is based on view of the recurrence of sub-atomic parts in truly realistic particles that relates without any difficulty of union. The fragmental commitment of SA is required to be ideal for successive substance moieties and troublesome for uncommon moieties (Table 3) [6].
| Assigned Mol-ID | Name of the compound | Similar Approved DRUG | % Similarity | Targets UniProt ID for FDA Approved Drug |
|---|---|---|---|---|
| 1A1.1 | Sitosterol | Allylestrenol | 91.2 | P06401 |
| 2A1.2 | Daucosterol | Ouabain | 59.8 | P05023 |
| 3A1.3 | Fucostanol | Menthol | 92.9 | Q7Z2W7 |
| 4A1.4 | β-Sitosterol acetate | Nandrolone decanoate | 71.4 | P10275 |
| 5A1.5 | Clionasterol | Allylestrenol | 91.2 | P06401 |
| 6A2.1 | (2R,3S,4S)- leucocyanidin |
Hesperetin | 72.4 | P35610 |
| 7A2.2 | Leucocianidol | Hesperetin | 72.4 | P35611 |
| 8A3.1 | Syringin | Cefixime | 81.2 | P44469 |
| 9A4.1 | Quercetin | Niclosamide | 70.8 | P11712 |
| 10A5.1 | DL- Tryptophan |
L-Tryptophan | 100 | Q9UGM6 |
| 11A5.2 | L-5-hydroxy- Tryptophan | Oxitriptan | 100 | Q9RVD6 |
| 12A5.3 | DL-N- Acetyltryptoph an |
L-Tryptophan | 80 | Q9UGM6 |
| 13A6.1 | Serotonin | Melatonin | 75.9 | P48039 |
| 14A6.2 | Bufotenine | Melatonin | 81.9 | P48040 |
| 15A7.1 | Isochroman | Tioconazole | 74.1 | P10613 |
| 16A7.2 | 1,2,3,3a- Tetrahydropyr rolo[1,2- a]quinoxalin- 4(5H)-one |
Mepivacaine | 61.5 | Q9Y5Y9 |
| 17A7.3 | 7-Hydroxy-5- methoxy-2- phenyl- chromen-; 4- one | Cromoglicic acid | 57.9 | Q12791 |
| 18A7.4 | 5-(4-Chloro- benzylidene)- 3-ethyl-2- thioxo- thiazolidin-4- one | Pyridostigmin e | 84.3 | P22303 |
| 19A7.5 | 4-oxo- nonenal | Progesterone | 72.2 | P06401 |
Table 3. List of bioactive compounds from Musa their percentage of similarity with FDA approved drugs and drug targets.
In Table 3 the compounds selected are then searched in the Protein Data Bank to obtain the protein ID, template PDB-ID,
% similarity and protein class. The obtained template of the protein is further used to obtain the target protein for the molecular docking studies and this in turn helps us to obtain the % similarity of the protein and the compound and also provides the protein class and significance of the protein.
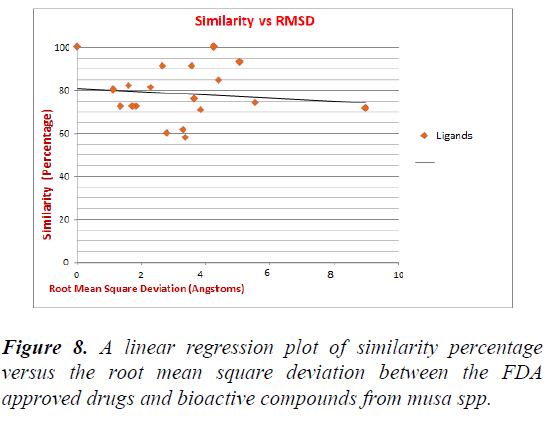
Further in Table the obtained template protein is then searched in the FDA approved drugs list to obtain the approved drug which is similar to that of our bioactive compound which gives the % similarity between the compound along with the calculation of Root Mean Square Deviation Value between Bioactive compounds and the FDA approved drug showed some significant RMSD Values. A Linear Regression Plot of Similarity Percentage versus the Root Mean Square Deviation between the FDA approved Drugs and Bioactive Compounds from Musa spp. Reveled the Significant Value of R2=0.0131 (y=-0.7202x+80.805) upon which approximately 6 Ligand molecules fall near on the regression line. The compound with more than 75% similarity and less RMSD values are considered for the further process of drug design; since they are very similar to the approved FDA drug they are most likely to show the ability to be a good lead molecule in the drug development and production (Figure 8) [7].
Figure 8: A linear regression plot of similarity percentage versus the root mean square deviation between the FDA approved drugs and bioactive compounds from Musa spp.
Future scope
Molecular docking evaluation administered with different compound which are a part of Phyto-constituents of Musa species which can fulfill the Pharmacological parameters considered under the studies will provide a robust information for in-vivo studies of those evaluated Phyto-constituents will guide them to be LEAD molecules which can grow into LIGAND for a possible DRUG molecules from Musa species for maximum Health benefits.
Conclusion
Musa is one among the a few genera inside the family Musa- ceae. Musa is a monoecious herb. The part of plantain like leaves, fruits, peels, roots are commonly utilized in the treatment of Snake bite, Hypertension, Antidepressant, Diabetes, Asthma, Insomnia, and Anthelmintic. This plant is documented for its many biological activities. Many scientists have worked with the plant sample and proved the presence of phytochemicals properties present within the plant. Several epidemiological and test contemplates have exhibited the numerous organic exercises of plantain. The fruits of this plants consists of starch, sugars, amino acids, vitamins and carbohydrates and therefore the flowers are known to possess secondary metabolites like sterols, saponins, tannins, non- reducing sugars and reducing sugars.
The compounds present within the plant are identified and analogs are identified for the further drug development process. The obtained analogs are then studied to understand the physicochemical properties like relative molecular mass, number of atoms, rotatable bonds count, molar refractivity, TPSA, and lipophilicity value, water solubility rate and sophistication, pharmacokinetics properties of the compound, drug-likeness properties like Lipinski, Ghose, Veber and Bioavailability score is additionally studied. And in medicinal chemistry lead-likeness and artificial accessibility is studied to understand whether the compound is eligible to become a lead molecule in drug formulation and to be an oral drug candidate.
After the study of physicochemical properties the compounds which are valid to be lead molecule are considered for next step to spot the target protein. Therefore the compounds are searched within the FDA approved drug data bank to get the compounds almost like the chosen compound then their target protein is identified to conduct the molecular docking studies to obtain the stable complex of molecule and protein binding.
Molecular docking evaluation administered with different compounds which are a part of Phytoconstituents of Musa species which have shown to fulfill the Pharmacological parameters considered under the studies will provide a robust information for in-vivo studies of those evaluated Phytoconstituents and guide them to be LEAD molecules which are growing LIGAND for a possible DRUG molecules from Musa species.
References
- Swathi D, Jyothi B, Sravanthi C. A review: pharmacognostic studies and pharmacological actions of Musa Paradisiaca. Int J Inn Pharm Res. 2011;2:122-5.
- Ittiyavirah S, Anurenj DA. Adaptogenic studies of acetone extract of Musa paradisiaca L. fruit peels in albino Wistar rats. International Journal of Nutrition, Pharmacology, Neurological Diseases. 2014;4:88.
- Ferreira LG, Dos Santos RN, Oliva G, Andricopulo AD. Molecular docking and structure-based drug design strategies. Molecules. 2015 ;20:13384-13421.
- Venkatesh KV, Girish KK, Pradeepa K, et al. Antibacterial activity of ethanol extract of Musa paradisiaca cv. Puttabale and Musa acuminate cv. grand naine. Asian J Pharm Clin Res. 2013;6:169-72.
- Rao MS, Gupta R, Liguori MJ, et al. Novel computational approach to predict off-target interactions for small molecules. Frontiers in big data. 2019;2:1-25.
- Jorgensen WL, Duffy EM. Prediction of drug solubility from structure. Advanced drug delivery reviews. 2002;54:355-66.
- Anandakumar S, Kannan D, Wilson E, et al. Potential phytopharmaceutical constituents of Solanum trilobatum L. as significant inhibitors against COVID-19: Robust-binding mode of inhibition by molecular docking, PASS-aid bioactivity and ADMET investigations. Biol Med Chem. 2020;2: 56-78.