Research Article - Biomedical Research (2017) Volume 28, Issue 18
Information retrieval for biomedicine applications through linked open data based optimization model
Senthilselvan N*, Subramaniyaswamy V and Sekar KR
School of Computing, Sastra University, Thanjavur, India
Accepted date: August 17, 2017
Abstract
In biomedical, objective of the semantic web is a pillar to connect plenty number of related data for information. Human and machine readable is the main theme behind web of science. Open linked data is the guideline for merging concepts, location and persons. Existing work is employed on Resource Description Framework (RDF) for machine learning purpose, GRDLL, POWDER, R2RML and SHACL are the existing work available in the market. The limitations of RDF for manipulating the data is not providing intellectual information retrieval, SPARQL is the query language playing a vital role. RDF is a lightweight and not possible for creating negative ontology. It is not holds good for negative expressions, cardinal values and metadata. The proposed methodology consisting of try model: (i) Backward chaining methodology in artificial intelligence; (ii) OWL-RDF is an intellectual query retrieval and (iii) Open linked data is used for obtain information’s from heterogeneous sources. It concludes Web Ontology Language (OWL) is the logic based language with a fine interoperability in web contents and always expresses the clear meaning. Using different biomedical datasets HCUP, data.gov, healthdata.gov, HMD, SEER, MHOS, DRUGBASE, and so on, open linked data consume information through the technique called OWL choreography.
Keywords
Biomedical applications, Linked open data, Backward chaining, Resource description framework (RDF), Web ontology
Introduction
In biomedical, biology and medical information combined together to provide the knowledge towards human life style. Analyzing, diagnosing and treatment are to be prescribed to the patient for their good hale and health. Information can be obtained from the confederation as data science. Open linked data is a phenomenon playing an important role in collecting information from different resources. Different web pages got their own significance in providing unsurpassed information. In open linked data inter and intra topic search will improve 14%. Personalized page ranking methodology was imposed in terms with preference and keyword based search [1]. Every nuke and corner of the information should be grabbed without leaving any stone unturned in the web application. Information’s to be linked together using the web pages and fetch via OWL-RDF is referred to as web of science. Service oriented architecture is implemented for open linked data to fetch the necessary data with a very great accuracy. Efficiency can be improved through ontology. Interoperability and reuse can be possible through the web content ontology. Ontology removes ambiguity in CDSS and appreciated in distributed system [2].
Semantic model, ontology and taxonomy are produced a greater inference. Meaning relation set of knowledge and good classification needs to be incorporated before arriving with a good result. According to the query, retrieving information can be made through linked open data in an optimized way with utility based reflex. Millions of tuples and hundreds of columns in an entity to be processed quickly through the semantic web rather OWL RDF to OWL RDF communication. RDF has got much significance in exchanging the data across the web pages. Linked open data now in the area of epidemiological studies. Medical data has got legal constraint and ethical measures. The aggregated data should need some ethical dispense for providing accurate results to the patient conditions. Three models were imposed as meta, profile and policies [3]. It is possible to integrate different protocols, different structures through RDF. If the schema changes at that time also possibility is more in merging structures. Semantic web is used to exchange, share and reuse the data across enterprise applications. Sentiment analysis and opinion mining are the subsets of polarity classification. In recent year’s natural language processing increases greater attention in the above said. Polarity classification brings up an ‘N’ number of patterns that exhibits polar facts. 79.98 percent precision can be found through the above said methodology [4,5].
Linked open data is well known for structured data and provides standard results in diversified web content pages. Semantic linked open data provides data through SPARQL that can be shared for ‘N’ number of web pages. Here the methodology applied was semantic similarity between resources and it is used for good recommender systems [6]. SPARQL is used for obtaining knowledge from heterogeneous application. SQL end points provide efficient result in gathering information. Optimizing the result can also be possible via SPARQL and it can always refer the hierarchical ontology tree [7]. OWL RDF is a computational logic based language which has got a reasoning power with the description logic. OWL RDF overseeds RDF and RDFS through their light weight component. Contribution to the research work is summarized as nutshell.
Biomedical query often leads to other confused query to the search engines. In the research, our ultimate aim is to optimize the end users query to the correct form so that the people will get a right URL and correct information. The query optimization has been down via the backward chaining model through artificial intelligence. Compared with other research models, the backward chaining model provides greater performance. It is already well discussed in the results and discussions segment through the Table 1. Moreover OWLRDFs is the intellectual machine to machine reader that brings all right information to the users.
| Methodology | Applications | Performance |
|---|---|---|
| Backward chaining | Query knowledge engineering | 91.43 |
| Reasoning systems | ||
| Business engines | ||
| Optimistic chaining | Tele-communication | 91.03 |
| Decision support system | ||
| Constraint handling rules | Constraint programming | 89.95 |
| Multi-agent system | ||
| Natural language processing | ||
| Temporal-spatial reasoning | ||
| Depth first backward chaining | Logic programming | 89.67 |
| Control strategies | ||
| Heuristic search | ||
| Exhaustive search | ||
| Business rule engines | Runtime production environment | 89.45 |
| Business process management | ||
| Rete algorithm | Pattern matching | 89.04 |
| Multiple strategy selection |
Table 1. Performance evaluation.
Related Works
Medical side needs high safety for their patients. Drugs with is sold over the counter rather self-medication provides injury to the patients. Through the linked open data sets somewhat provide relevant information and characteristics about the present condition of the disease [4]. Web service facilitates greater functionality to the interface for collecting collective information and uniform retrieval towards bio information. OWL and OBO were helped in getting meta data [8]. Biomedical big data is the community oriented platform to encourage high quality of knowledge in the bio field. The framework encourages in sharing knowledge and personalization content [9]. In biomedical application selfhealing property is encouraged in terms with removing stiffness, toughness. Stiff, tough and self-healing are the major goals for the healing property. Self-healing has recorded 67 percent of original strength after 10 min once it reaches high serum level. Hydrogel research that holds high potential in the diversified biomedical application [10]. In biomedical data LOD provides greater integrity, query and visualization.
The scalability of data can be improved through federated query in the distributed web content data. User friendly interface can possibly improve the customer query through their questionnaire [11]. In biomedical research health care system has got a great demand and urgent solutions. Hypothesis driven methodology has been shipment to data driven approach. Efficient interpretation through big data is essential for physiological and pathological states. New paradigm accessing methodology and cognitive knowledge gives efficiency to the biomedical interfaces. High performance computing system needs to be incorporated for optimizing the solution retrieval [12].
Data interoperability provides a greater knowledge for potential interaction. Through open linked data susceptible data can be detected in the poly pharmacy application. Open linked data is also applicable for osteoarthritis, diabetes and hypertension application.
Conflict recommendations have to be detecting before finding the resultant [13]. RDF and SPARQL are the essential tools for retrieval data in natural language processing. Two methodologies provide extensive knowledge in the biomedical side. Linguistic approach and semantic annotation are the two step process appreciated for the cognitive knowledge [14].
In biomedical, pathology tells how disease caused and what type of drug that should be implemented for the above said. Ontology provides greater dissemination and knowledge that can be obtained through SPARQL [15].
Proposed Optimized Architectural Model
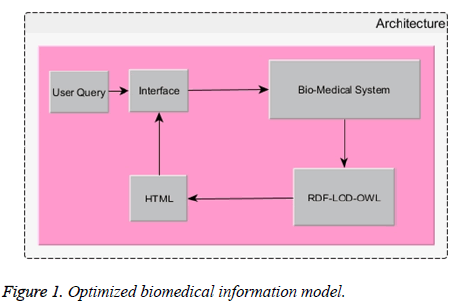
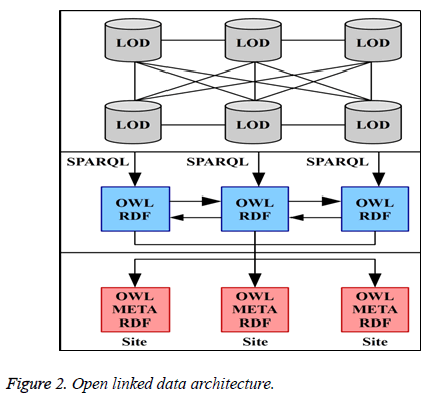
Figures 1 and 2 gives the detailed optimized model for retrieving biomedical data for the end users. The information retrieved through the optimized model provides greater precision in the information.
Biomedical queries first hit the user interface. The same query will be send back to bio-medical system. In this system backward chaining method is incorporated for refactor the user’s query to the correct form.
The query will be decomposed into tokens called atomic words. These atomic words are fed to backward chaining system. After ‘N’ iterations the formation of correct query attained at the top of the backward chaining system. The corrected or refactored query now sends to RDF system.
The intellectual OWL-RDF will collect all the necessary information from different resources with accurate measures. As we know OWL-RDF stands one step ahead of RDF and RDFS.
All relevant information’s are collected in html format. Open linked data principles were adapted while taking information for the query from heterogeneous domain rather as cognitive knowledge.
Figure 2 has detailed information about OWL RDF in the paradigm and the same is incorporated in the Figure 1. The outcome of the result provides 91.36% accuracy because of backward chaining and open linked data. The resultant html is send back to the user interface as a response. Here optimization has done through refactoring the query using backward chaining method. Moreover OWL-RDF is a meticulous query pattern capable of grapping at most right results. In this LOD based paradigm OWL-RDF and BCM optimized the model to the core. The model gets accelerated through their accuracy because of refactoring the query using a right system.
Ideology and inference of the optimized model
The model proposed to find the correct information for the query. The model optimizes with three great ideologies: (i) Educate the query through artificial intelligence BCM; (ii) OWL-RDF is an intellectual information grabber from the query; (iii) Applied open linked data for retrieval of information from multiple sources. Optimizing the model not only brings-up the data fetching in a right manner but also save the cost factor. Accuracy and performance efficiency can be seen deliberately through this model and it gives more mileage in the era.
Results and Discussions
Backward chaining model in artificial intelligence, OWL-RDF and open linked data were the tools employed to optimize the biomedical inference model for retrieving the correct URL and at most right answer for the query given by the end user. The cost factor is also involved in the work that gives added value greatness to the model is all proposed here. The open line of work in terms with biomedical data model optimization is the eye opener for the researchers in the years to come. Performance analysis methods are as follows.
Performance=(Ideal cycle time × Total count)/Run Time (1)
Ideal cycle time=It is all about arriving query into the system
Total count=Total number of query arrived
Run time=How much time it is taken to resolve the query to the correct sense.
Backward chaining model will improve and optimize the query to the greater extends to get the get the correct information for the bio medical data query. Large sample set of 1742 biomedical query was investigated and tested for performance evaluation using equation (1). Using the above said formula, performance measures have been calculated and the same is shown in the Table 1. In Table 1, relevant other researcher methods, applications and the performance are projected clearly.
Conclusion
Biomedical optimized model should be constructed in a way to produce correct information to the medical customer and in a cost effective manner. The tri-model methodologies involved in the work brings up the new era to the field of biomedical model optimization. Still many new ideas needed to be incorporated to generate effective results in terms of reduced time and space complexities. The above said measures should be pathway in the unforeseen schedule of our research.
References
- Zong N, Lee S, Ahn J, Kim HG. Supporting inter-topic entity search for biomedical linked data based on heterogeneous relationships. Comput Biol Med 2017; 87: 217-229.
- Marco-Ruiz L, Pedrinaci C, Maldonado JA, Panziera L, Chen R, Bellika JG. Publication, discovery and interoperability of clinical decision support systems: A linked data approach. J Biomed Inform 2016; 62: 243-264.
- Kamateri E, Kalampokis E, Tambouris E, Tarabanis K. The linked medical data access control framework. J Biomed Inform 2014; 50: 213-225.
- Abula T, Worku A. Self-medication in three towns of North West Ethiopia. Ethiopian J Health Development (EJHD) 2017; 15.
- Noferesti S, Shamsfard M. Using Linked Data for polarity classification of patients’ experiences. J Biomed Inform 2015; 57: 6-19.
- Meymandpour R, Davis JG. A semantic similarity measure for linked data: An information content-based approach. Knowledge-Based Systems 2016; 109: 276-293.
- Purawat S, Cowart C, Amaro RE, Altintas I. Biomedical Big Data Training Collaborative (BBDTC): An effort to bridge the talent gap in biomedical science and research. J Comput Sci 2017; 20: 205-214.
- Sabzi M, Samadi N, Abbasi F, Mahdavinia GR, Babaahmadi M. Bioinspired fully physically cross-linked double network hydrogels with a robust, tough and self-healing structure. Materials Sci Eng 2017; 74: 374-381.
- Saleem M, Kamdar MR, Iqbal A, Sampath S, Deus HF, Ngomo ACN. Big linked cancer data: Integrating linked tcga and pubmed. Web Semantics: Science, Services and Agents on the World Wide Web 2014; 27: 34-41.
- Merelli I, Pérez-Sánchez H, Gesing S, D’Agostino D. Managing, analysing, and integrating big data in medical bioinformatics: open problems and future perspectives. Biomed Res Int 2014.
- Zamborlini V, Hoekstra R, da Silveira M, Pruski C, ten Teije A, van Harmelen F. Generalizing the detection of clinical guideline interactions enhanced with LOD. In International Joint Conference on Biomedical Engineering Systems and Technologies: Springer, Cham 2016; 360-386.
- Hamon T, Grabar N, Mougin F. Querying biomedical linked data with natural language questions. Semantic Web 2016; 8: 581-599.
- Marginean A. Question answering over biomedical linked data with grammatical framework. Semantic Web 2017; 8: 565-580.
- Hasnain A, Mehmood Q, e Zainab SS, Saleem M, Warren C, Zehra D, Rebholz-Schuhmann D. BioFed: Federated query processing over life sciences linked open data. J Biomed Semantics 2017; 8: 13.
- Kim JD. Biomedical natural language processing. MIT Press J 2017; 43: 265-267.