Research Article - Biomedical Research (2018) Computational Life Sciences and Smarter Technological Advancement: Edition: II
In silico prediction of interacting domain of RIP1 with p38 MAPK
Rini Jacob and Anbalagan M*
School of Biosciences and Technology, VIT University, Vellore, India
- *Corresponding Author:
- Anbalagan M
School of Biosciences and Technology
VIT University, India
Email: anbalagan.m@vit.ac.in, Phone Number: 9489333231
Accepted on February 01, 2017
DOI: 10.4066/biomedicalresearch.29-16-2324
Visit for more related articles at Biomedical ResearchAbstract
Extracellular receptors convey specific signals to the nucleus for modulation of gene expression through signaling molecules present in the cytoplasm. RIP1 is a signaling molecule involved in the proximal end of TNFα signaling. Signal transduction pathways involving RIP1 and JNK/p38 MAPKs are essential in inflammation and hence this signaling pathway serve as a target for novel drug development in inflammation mediated disorders. Though, the role of RIP1 in p38 MAPK activation have been reported before, the exact role of this signaling molecule in p38 MAPK activation is not clear. In silico prediction of RIP1 binding with p38 MAPK suggests a domain with amino acids 71-79 is involved in the interaction. Deletion of 71-79 amino acids in RIP1 destabilizes the interaction with p38 MAPK. This prediction if confirmed with wet lab analysis, can serve as a target for designing novel drugs for treating various disorders.
Keywords
Protein- protein interactions, RIP1, p38 MAPK, Molecular docking.
Introduction
The cell is constantly exposed to various kinds of stimuli, both physical and chemical. A multicellular organism, respond to external stimuli by means of differential gene expression, where mRNA population and hence protein composition changes in the cell in such a way to respond to the stimuli. Differential gene expression is involved in various physiological processes from cell proliferation to cell death that are mediated by distinct and specific signal transduction mechanisms [1]. Extra cellular stimuli are conveyed to the nucleus for changes in gene expression through a biochemical reaction called as transduction process. The specificity among the different signaling pathways are brought about by differences in protein – protein interaction, protein-DNA interactions and protein-RNA interactions [2]. With respect to signaling, protein-protein interactions (PPI) play a major role in the cells. Different proteins present in the cell interact with other proteins using specific signature domains/motifs [3]. The amino acid sequences involved in these interactions are so important that a single change in the amino acid sequence of the proteins would lead to defective signaling, affecting the physiology of the cells/organism [4].
RIP1 (Receptor Interacting Protein kinase 1) belongs to the family of Receptor Interacting Protein (RIP) kinases, a group of serine/ threonine kinases [5]. They comprise of seven members (RIP1-RIP7) with a conserved and homologous kinase domain [6]. They are known to be involved in transducing various signaling cascades playing an important role in immune response, inflammation, and apoptosis. RIP 1 consists of a death domain, which when recruited initiates several signaling pathways and acts as a molecular switch between inflammation and apoptosis. RIP1/RIP3 is known to mediate necroptosis [7]. Earlier reports have suggested that RIP1 mediates p38 MAPK activation in response to TNFα through MEKK3 [8]. RIP1 plays a regulatory role in activating the MAPKs. RIP1 is an essential modulator of TNFα induced necrotic cell death [9], since RIP1 is known to mediate apoptosis and necrotic cell death, the involvement of these kinases in various pathological conditions is crucial. RIP1 is linked to various diseased conditions including heart attacks, pancreatitis, liver, retina and renal injury [10]. Given its importance in various signaling processes, a detailed study on molecular mechanism of RIP1 signaling in cell will lead to development of novel drugs to treat physiological conditions related to the malformation of RIP1 signaling [11].
Mitogen activated protein kinases (MAPK) are a group of evolutionarily conserved serine/threonine protein kinases, which acts as a central point for mediating a broad range of stimuli to their respective transcription factors, which in turn modulate expression of relevant genes required to respond to physiological situation [12]. The major MAPKs include ERK1/2, JNK1/2 and p38 MAPK. JNK1/2 and p38 MAPK are known as Stress Activated Protein Kinases (SAPKs) due to their response against various stress factors such as UV, osmotic radiation, hypoxia and inflammatory cytokines [13]. p38 MAPK is a 38kDa protein with four different isoforms α, β, γ and δ. Among the four, p38α (MAPK14) is ubiquitously expressed while the other isoforms are expressed tissue specifically. The canonical activation of p38 MAPK is by the dual phosphorylation of threonine and tyrosine of the TGY motif at the positions 180 and 182 respectively by the upstream kinases MKK3/6 [14]. The activated p38 MAPK further activates various downstream signaling molecules such as kinases, phosphatases and transcription factors leading to various physiological functions. Recent studies have also shown the involvement of p38 MAPK in various cancers, cardiovascular dysfunction and Alzheimer’s disease [15]. Understanding the interacting partners of p38 MAPK would give us an insight into its molecular aspect of signaling. We have used in silico tools to predict a model for RIP1 and MAPK complex and predicted the domains involved in the interaction of RIP1 with MAPK.
Materials and Methods
Preparation of protein molecules
The initial docking protocol includes the retrieval of x-ray crystal structure of the p38MAPK and RIP1 protein. The protein structure of p38MAPK and RIP1 protein was obtained from Protein Databank with PDB IDs 1A9U and 4ITJ [16] respectively. The protein structure was energy minimized before using SWISS PDB Viewer [17] to avoid any false predictions. The energy minimized structures of the proteins were then subjected to further docking protocol.
Homology modelling
Homology modeling of the RIP1 protein without the D-motif was performed using Swiss-Model [18]. The sequence of the protein was retrieved from UniProt database with ID: Q13546. The homology modeling of the protein was performed with the same structure of RIP1 with PDB ID 4ITJ (16) was used as a template in molecule preparation.
Molecular docking
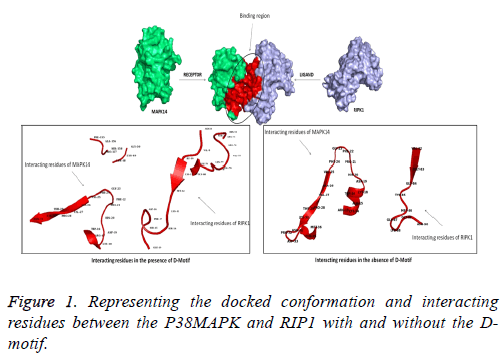
The protein-protein docking was performed with the help of HADDOCK [19], ZDOCK [20] and ClusPro [21]. Since the p38MAPK are highly specific by interacting with the D-motif present in the interaction protein, we have identified all the Dmotif regions present in RIP1 and was given as the binding/ active site in HADDOCK and Z-Dock, whereas a blind docking analysis was performed in the case of ClusPro. Also to have better comparative analysis, and understand the binding with and without the D-motif, the protein model without Dmotif was also docked with p38MAPK. The interacting region and residues were visualized with the help of PyMOL.
Results
Molecular docking analysis
The interaction and interacting residues between the P38MAPK and RIP1 proteins were analyzed with the aid of molecular docking analysis. As a result of docking, we obtained many docked poses of which, the best-docked pose were selected based on the least binding energy, least RMSD score and interaction of P38MAPK with the D-motif present in the RIP1 protein. Primarily, the docked poses with high binding energy were excluded and further any docked pose with more than 1.0 Å RMSD were also excluded from the study. Finally, the remaining docked complexes were analyzed for interaction with the D-motif residues. The best ten models were taken for further screening.
The binding energy and RMSD values of each docked poses are tabulated in Tables 1 and 2. While screening the docked poses, we identified that one of the docked poses; P38MAPK interacted with Arg71, His72, Ser73, Arg74, Val75, Val76, Lys77, Leu78 and Leu79 of RIPK1 which satisfies the formula of the D-motif. Along with these residues, we also found many other interacting residues in RIP1 and P38MAPK which are tabulated in Table 3. Although interacting residues are important, more critical are the various interactions such as van der Waal’s forces, electrostatic interactions. The best-docked pose was analyzed for the above-stated interactions (Table 5). When the docking was performed without the D-motif in the RIP1 protein, we found lesser interactions between the RIP1 and P38MAPK. The interacting residues of the proteins are tabulated in Table 4. Also, we found lesser electrostatic energy observed between the proteins (Figure 1 and Table 5).
| Model Number | Binding energy kcal/mol | RMSD (Å) |
|---|---|---|
| Model1 | -128.65 | 0.87 |
| Model2 | -123.73 | 0.93 |
| Model3 | -109.8 | 1.12 |
| Model4 | -112.96 | 0.98 |
| Model5 | -118. 43 | 0.900 |
| Model6 | -121.46 | 0.991 |
| Model7 | -113.43 | 1.110 |
| Model8 | -127.77 | 0.98 |
| Model9 | -127.37 | 0.921 |
| Model10 | -122.33 | 1.112 |
Table 1. The best ten docked poses with its binding energy and RMSD values between p38 MAPK and RIP1 with D-motif.
| Model Number | Binding energy kcal/mol | RMSD (Å) |
|---|---|---|
| Model1 | -113.66 | 0.895 |
| Model2 | -110.87 | 0.92 |
| Model3 | -112.63 | 1.005 |
| Model4 | -103.67 | 0.924 |
| Model5 | -109.44 | 1.015 |
| Model6 | -113.34 | 0.966 |
| Model7 | -121.33 | 0.897 |
| Model8 | -118.44 | 0.904 |
| Model9 | -114.22 | 0.887 |
| Model10 | -113.34 | 0.945 |
Table 2. The best ten docked poses with its binding energy and RMSD values between p38 MAPK and RIP1 without D-motif.
| Proteins | P38MAPK | RIP1 |
|---|---|---|
| Interacting Residues | Trp16, Arg17, Lys18, Asp19, Pro21, Phe22, Gly23, Phe24 Val25, Ala26, Val27, Pro28, Thr29, Lys30, Asn31, Lys48, Gly50, Phe155, Ala156, Pro157, Ser158 | An8, Val9, Ile10, Lys11, Let12, Lys13, Ser14, Ser15, Asp16, Phe17, Glu19, Arg71, His72, Ser73, Arg74, Val75, Val76, Lys77, Leu78, Leu79 |
Table 3. The interacting residues of P38MAPK and RIP1 with Dmotif.
| Proteins | P38MAPK | RIP1 (Without D-motif) |
|---|---|---|
| Interacting Residues | Arg 13, Lys14, Ala15, Trp16, Lys18, Asp19, Pro21, Phe22, Gly23, Phe24 Val25, Ala26, Val27, Pro28, Thr29, Lys30, Asn31, Pro32, Asp33, Thr 35, Met36 | Val82, Met83, Glu84, Tyr85, Met86, Glu87, Gly89, Asn90 |
Table 4. The interacting residues of P38MAPK and RIP1 without Dmotif.
| Protein-protein Complex | Binding Score | Electrostatic potential | Van der Waal’s energy | Buried surface Area |
|---|---|---|---|---|
| P38MAPK-RIP1 | -127.37 | −433.0 ± 16 | −144.2 ± 11 | 3997.3 ± 82.4 |
| P38MAPK-RIP1 (Without D-motif) | -121.33 | -419.3 ± 12 | −132.78 ± 10 | 3994.8 ± 78.6 |
Table 5. Interactions observed in the protein- protein complex as results obtained from HADDOCK.
Discussion
Identification of protein - protein interactions that occur in the cells is of interest in biology so that unique protein-protein interaction mediating a physiological function can serve as target for design of novel drugs [22]. Inhibitors of p38 MAPK are sought in medical field as an alternate for treating inflammatory diseases [23]. MAPKs are important signaling molecules playing a central role of conveying messages from extra cellular receptors to transcription factors through upstream kinases [12,13]. Inhibition of MAPKs is not the good choice for treatment of a particular disease, for the reason that inhibition of MAPKs in a cell would affect other functions in a cell/organism, leading to side effects. Currently, signaling pathways and signaling molecules involved in a particular pathway mediating specific function of the cell need be identified so that drugs affecting specific molecules/pathways can be developed that would expect to have less side effect.
Since protein – protein interactions play an important role in signal transduction pathway, and considering the importance of p38 MAPK, our lab is interested in identification of proteins that interact with p38 MAPK. RIP1-p38MAPK interaction is of interest to us for the reason that both molecules are involved in mediating TNFα signaling [24]. Role for RIP1 mediated p38 MAPK activation is not clear [24-26]. Upon TNFα stimulation, RIP is known to get autophosphorylated and activate downstream signaling molecules including p38 MAPK leading to cellular death [9,27]. TNFα was not able to activate p38 MAPK in RIP 1-/- fibroblast, however when KD mutant of RIP1 was expressed in these cells, TNFα mediated p38 MAPK activation was observed, suggesting kinase activity of RIP1 or this domain are not required for p38 MAPK activation [24]. All the MAPK interacting proteins use a signature motif, known as D motif (- (R/K)1-2 – (X) 2-6 – L/I/V – X- L/I/V-) to interact with MAPKs [28]. A search for this motif in the amino acid sequence revealed three D motifs (71RHSRVVKL; 190KPENILV; 203KIADLGL). In silico approach was taken to study molecular interaction between p38 MAPK and RIP1. Molecular docking protocols offer insights over the fundamental interactions in the protein and between the proteins [29]. The docking calculations involving RIP1 and p38 MAPK, obtained from three servers yielded about 30 different docked conformations. Of the three servers, the results from ClusPro were eliminated as primarily, it was a blind docking analysis, and also none of the poses showed a whole interaction with the D-motif. The docked solutions of ZDOCK and HADDOCK were further analyzed in the current study. The best-docked poses were screened with the help of least binding energy and showing interaction with the D-motif present in the interacting protein (in RIP1). This docked conformation were then subjected to other interaction analysis such as van der Waals and electrostatic potentials as these are a few significant interaction [30,31]. Similarly, when comparing the various interactions, we found there were electrostatic energy and van der Waals forces observed in the proteinprotein complex which may further aid in stronger interactions. When analyzing the binding patterns without the D-motif, we found lesser residues from both the protein to be involved interaction and also we found there was a decrease in electrostatic energy and van der Waals forces. These results help us to understand the fact that, the P38MAPK interacts with this possible part in the RIP1 protein.
Inhibitors for RIP1 have been synthesized; however these inhibitors are known to have several side effects [11,32]. RIP1 being upstream kinase, several pathways may be regulated by RIP1 hence blocking RIP1 function would block other crucial signal transduction pathways leading to side effects [33]. Though in vitro phosporylation of p38 MAPK by RIP1 have been reported earlier [24] there are no report available about p38 MAPK physical interaction, here we present evidences for interaction between these molecules from in silico analysis. More studies are required to carry out for understanding the physiological significance of this interaction and signal transduction pathways/signaling molecules regulated so that novel drugs that target single signal transduction pathway can be developed that will have less side effects.
Acknowledgement
We gratefully acknowledge Department of Biotechnology, Govt. of India, for providing financial support through the sponsored project titled “Identification of novel p38MAPK interacting proteins involved in inflammation mediated cell death”.
References
- Lodish H, Berk A, Zipursky S L, Matsudaira P, Baltimore D, Darnell J. Molecular Cell Biology. 4th ed. New York: W. H. Freeman and Company, 2000.
- Hornák V, Dvorský R, Sturdík E. Receptor-ligand interaction and molecular modelling. Gen Physiol Biophys 1999; 18: 231-248.
- Janin J, Wodak SJ. Structural domains in proteins and their role in the dynamics of protein function. Prog Biophys Mol Biol 1983; 42: 21-78.
- Tony P, Nash P. Protein–protein interactions define specificity in signal transduction. Genes Dev 2000; 14: 1027-1047.
- Stanger BZ, Leder P, Lee TH, Kim E, Seed B. RIP: a novel protein containing a death domain that interacts with Fas/APO-1 (CD95) in yeast and causes cell death. Cell 1995; 81: 513-523.
- Zhang D, Lin J, Han J. Receptor-interacting protein (RIP) kinase family. Cell Mol Immunol 2010; 7: 243-249.
- Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, Damko E, Moquin D, Walz T, McDermott A, Chan FK, Wu H. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 2012; 150: 339-350.
- Yang J, Lin Y, Guo Z, Cheng J, Huang J, Deng L, Liao W, Chen Z, Liu Z, Su B. The essential role of MEKK3 in TNF-induced NF-kappaB activation. Nat Immunol 2001; 2: 620-624.
- Holler N, Zaru R, Micheau O, Thome M. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nature Immunol 2000; 1: 489-495.
- Christofferson DE, Li Y, Yuan J. Control of life-or-death decisions by RIP1 kinase. Annu Rev Physiol 2014; 76: 129-150.
- Degterev A, Hitomi J, Germscheid M, Ch'en IL, Korkina O, Teng X, Abbott D, Cuny GD, Yuan C, Wagner G, Hedrick SM, Gerber SA, Lugovskoy A, Yuan J. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol 2008; 4: 313-321.
- Karin M. Mitogen-activated protein kinase cascades as regulators of stress responses. Ann NY Acad Sci 1998; 851: 139-146.
- Davis RJ. Signal transduction by the c-Jun N-terminal kinase. Biochem Soc Symp 1999; 64: 1-12.
- De´rijard B, Raingeaud J, Barrett T, Wu IH, Han J, Ulevitch RJ, Davis RJ. Independent human MAP kinase signal transduction pathways defined by MEK and MKK isoforms. Science 1995; 267: 682-685.
- Ana C, Rousseau S. p38 MAP-Kinases pathway regulation, function and role in human diseases. Biochimica et Biophysica Acta 2007; 1773: 1358-1375.
- Xie T, Peng W, Liu Y, Yan C, Maki J, Degterev A, Yuan J, Shi Y. Structural basis of RIP1 inhibition by necrostatins. Structure 2013; 21: 493-499.
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 1997; 18: 2714-2723.
- Biasini M, Bienert S, Waterhouse A. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 2014; 42: W252-258.
- Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc 2003; 125: 1731-1737.
- Pierce BG, Wiehe K, Hwang H. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics 2014; 30: 1771-1773.
- Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res 2004; 32: W96-99.
- Jacob R, Anbalagan M. Targeting secret handshakes of biological processes for novel drug development. Front Biol 2016; 11: 132-140.
- Yong HY, Koh MS, Moon A. The p38 MAPK inhibitors for the treatment of inflammatory diseases and cancer. Expert Opin Investig Drugs 2009; 18: 1893-1905.
- Lee TH, Shank J, Cusson N, Kelliher MA. The Kinase Activity of Rip1 Is Not Required for Tumor Necrosis Factor-a- induced I?B Kinase or p38 MAP Kinase Activation or for the Ubiquitination of Rip1 by Traf2. J Biol Chem 2004; 279: 33185-33191.
- Anne B, Lin Y, Liu ZG. The role of the death-domain kinase RIP in tumor-necrosis-factor-induced activation of mitogen-activated protein kinases. EMBO Rep 2003; 4: 623-627.
- Kaamar A, Yuvaraj S, van-Roosmalen I. MAPK p38 and JNK have opposing activities on TRAIL-induced apoptosis activation in NSCLC H460 cells that involves RIP1 and caspase-8 and is mediated by Mcl-. Apoptosis 2013; 18: 851-860.
- Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 2003; 114: 181-190.
- Takuji T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat Cell Biol 2000; 2: 110-116.
- Vakser IA. Protein-protein docking: from interaction to interactome. Biophys J 2014; 107: 1785-1793.
- Smith GR, Sternberg MJ. Prediction of protein-protein interactions by docking methods. Curr Opin Struct Biol 2002; 12: 28-35.
- Thirumal Kumar D, George Priya Doss C, Sneha P, Tayubi IA, Siva R, Chakraborty C, Magesh R. Influence of V54M mutation in giant muscle protein titin: a computational screening and molecular dynamics approach. J Biomol Struct Dyn 2016.
- Harris Philip A, Bandyopadhyay D, Berger SB. Discovery of Small Molecule RIP1 Kinase Inhibitors for the Treatment of Pathologies Associated with Necroptosis. ACS Med Chem Lett 2013; 4: 1238-1243.
- Sakthivel K, Jayanthiladevi A , Kavitha C. Automatic detection of lung cancer nodules by employing intelligent fuzzy cmeans and support vector machine. Biomed Res 2016; 27: S123-S127.