Research Article - Journal of RNA and Genomics (2019) Volume 15, Issue 1
In silico Identification of Riboswitches in the Human Gut Microbiome for Therapeutic Applications
Priyanka Kumari and Anup Som*Centre of Bioinformatics, Institute of Interdisciplinary Studies, University of Allahabad, Prayagraj, UP, India
- *Corresponding Author:
- Anup Som
Centre of Bioinformatics
Institute of Interdisciplinary Studies
University of Allahabad, Prayagraj, UP, India
Tel: +91-9838115336
E-mail: som.anup@gmail.com
Received Date: 30 July 2019; Accepted Date: 05 October 2019; Published Date: 12 October 2019
Copyright The Author(s). Priyanka Kumari and Anup Som. First Published by Allied Academies. This is an open access article, published under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0). This license permits non-commercial use, distribution and reproduction of the article, provided the original work is appropriately acknowledged with correct citation details
Abstract
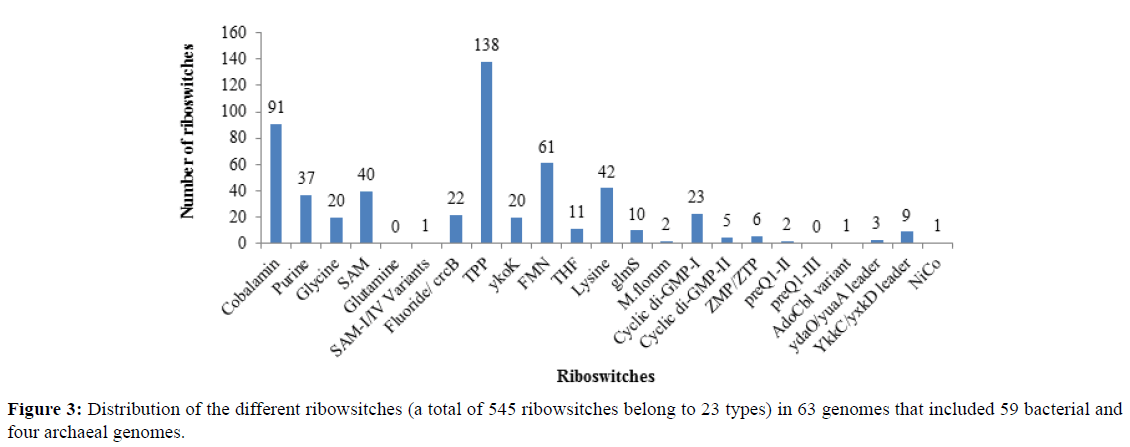
Riboswitches are cis-acting, folded non-coding RNA structures which regulate gene expression via conformational changes on binding with specific metabolite. They regulate gene expression of bacteria, archaea, fungi and some plants and may act as potent drug targets. The inhibition of the riboswitches controlling vital genes results in the suppression of growth or death of the organism. Since the normal human gut microbiome comprises of bacterial and archaeal colonies, the inhibition of riboswitches may lead to major changes in the composition of gut microbiome causing diseased condition in human. Therefore, in this study the distribution of various riboswitches, the genes regulated by them and their potential as RNA drug target was explored. The study identified 545 candidate riboswitches in 59 bacterial and 4 archaeal genomes of the adult human gut. The study also revealed that the most abundant riboswitch is the TPP riboswitch (25%) followed by Cobalamin (17%), FMN (11%) and Lysine riboswitch (8%). The lower abundance was shown by YkkC/yxkD leader (2%), Cyclic di-GMP II (1%) and ZMP/ZTP riboswitch (1%); the rare ones included M. Florum (0.4%), Nico (0.2%), AdoCbl variant (0.2%) and SAM-I/IV variant riboswitch (0.2%). Further, the genes regulated by these riboswitches were identified and seven riboswitches such as c-di-GMP I, c-di-GMP II, SAM, glmS, THF, YdaO/ YuaA leader, and glycine riboswitches were predicted as drug targets in the pathogenic bacteria of the human gut.
Keywords
Human gut microbiome, Riboswitch, Regulated genes, GC content, Drug target, Chemical analogs, Computational approach
Abbreviations
SAM: S-Adenosyl Methionine; TPP: Thiamine Pyrophosphate; FMN: Flavin Mononucleotide; THF: Tetrahydroflorate; GlmS: Glucosamine-6-phosphate; C-di-GMP: Cyclic Diguanylate
Introduction
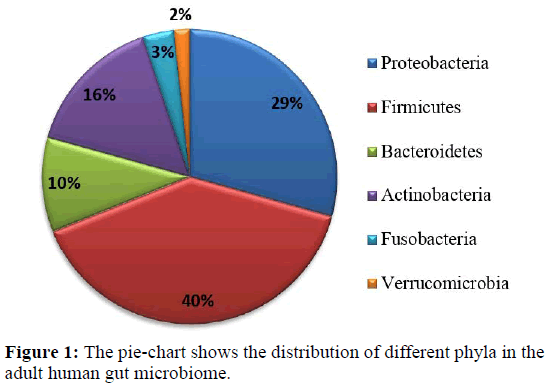
The human body contains a plethora of microbial organisms in their vicinity as metagenomic content. The human gut harbours complex population of different kind of microorganisms like bacteria, archaea and eukaryotes. Here the bacterial cells are ten times the number of human cells and genes counts are ten folds higher than the total gene count of human [1,2]. Generally, the normal adult human gut microbiota comprises of four major bacterial phyla namely Actinobacteria, Bacteroidetes, Firmicutes and Proteobacteria [3, 4]. The phyla Fusobacteria and Verrucomicrobia are present in lower proportions [5,6]. Greater variations exist below the phylum level, although certain butyrate-producing bacteria, including Bacteroides uniformis, Faecalibacterium prausnitzii, and Roseburia intestinalis have been identified as key members of the adult gut microbiota [7,8,9]. The composition may vary due to different diets, habitats, environment etc. [10,11]. The disturbance in the composition of microbial communities might cause an immune response from the commensal native bacteria in the gastrointestinal tract [1, 2]. The microbes in human gut influence human physiology, metabolism, nutrition and immune function so disruption of the gut microbiota is linked with various diseased conditions [3, 4, 11]. Thus major changes in the colonisation affect human health adversely and it’s prolong persistent may cause serious health issues.
Riboswitches are structured non-coding RNA elements present at the 5’ untranslated region (5’ UTR) of the mRNA which bind selectively to specific metabolite to regulate the adjacent gene expression. They are widespread in bacteria and have been also found in archaea, fungi and plants [13,14]. Riboswitches consist of a highly conserved aptameric region and a less conserved platform region and regulate gene expression at the transcriptional and post- transcriptional level [15, 16]. The aptamer binds selectively to specific metabolite/ ligand to induce conformational changes in the platform region which further regulates the adjacent gene [17, 18]. Therefore the analogs of this metabolite may bind to the riboswitches and affect the regulation of the fundamental metabolic pathways adversely [19, 20]. For example: Roseoflavin, an analog binds directly to the aptamer domain of FMN riboswitch declining the expression of a reporter gene located downstream of an FMN riboswitch in B. subtilis [21,22,23]. Pyrithiamine is pyrophosphorylated and then binds to TPP riboswitch [24]. L Aminoethylcysteine (AEC) and DL-4-oxalysine are lysine derivatives which reduce the growth of some gram-positive bacteria binding to lysine riboswitch etc. [25]. Thus riboswitches can be exploited as RNA-based biosensors and RNA-based drug targets [26,27,28]. These analogs may also fall in the category of drugs that we use. This is more important in case of oral medicines as they affect the human gut directly so it must be taken in consideration as it is done for the ADME (Absorption, Distribution, Metabolism, Excretion and Toxicity) properties, because if it is ignored then in long run they may remove a colony of microorganisms critical to health of the person which may cause disease in future.
Recently, in the drug target identification pipeline for microbes causing disease the human gut microbiome proteome is also taken into account [29,30]. Similarly for RNA-based drugs also the same should be considered. Hence, a deeper insight into flora of human gut especially in terms of the distribution of riboswitches (RNA biomolecules) is essential.
In this paper, the riboswitches in human gut microbiota had been identified and their distribution had been studied. The genes regulated by these riboswitches were identified and seven riboswitches were uncovered as potent RNA drug targets for gut bacterial pathogens. Further GC% of the genomes and riboswitches present in the corresponding genomes was also analysed.
Materials and Methods
Collection of microbial genome sequences
An extensive literature mining and information from NIH Human Microbiome Project led to the selection of genomes which represent bacterial and archaeal microflora of human adult gut [31]. The healthy human gut is represented mostly by the bacterial species from four major phyla firmicutes, bacteroidetes, actinobacteria and proteobacteria constituting 90% of the human gut microbiome [1,32]. The phyla Fusobacteria and Verrucomicrobia are present in lower proportions [5,6]. Recent studies reported that for understanding the homeostasis of the microbiome both the dominant and rare microbial community is taken into account [33,34]. Considering the above facts 63 whole genome sequences were retrieved and used for this study that included 59 bacterial and four archaeal species representing the minimal gut genome of adult microbiome (details are given in Supplementary Table S1) [31]. Figure 1 shows the phyla distribution of these different microbes. Out of 59 bacterial species 23 are pathogenic and 36 are non-pathogenic bacteria. The genome sequences were retrieved from NCBI microbial genome database (https://www.ncbi.nlm.nih.gov/ genome/microbes/). Among the 42 types of riboswitches reported in the Rfam database 23 types were considered as their 3-dimensional experimentally validated structures are present in the PDB database. The sequences of 23 types of riboswitches (listed in Supplementary Table S2) were taken as multiple sequence alignment file (Stockholm file) from Rfam database for the analysis [35].
Prediction of riboswitches
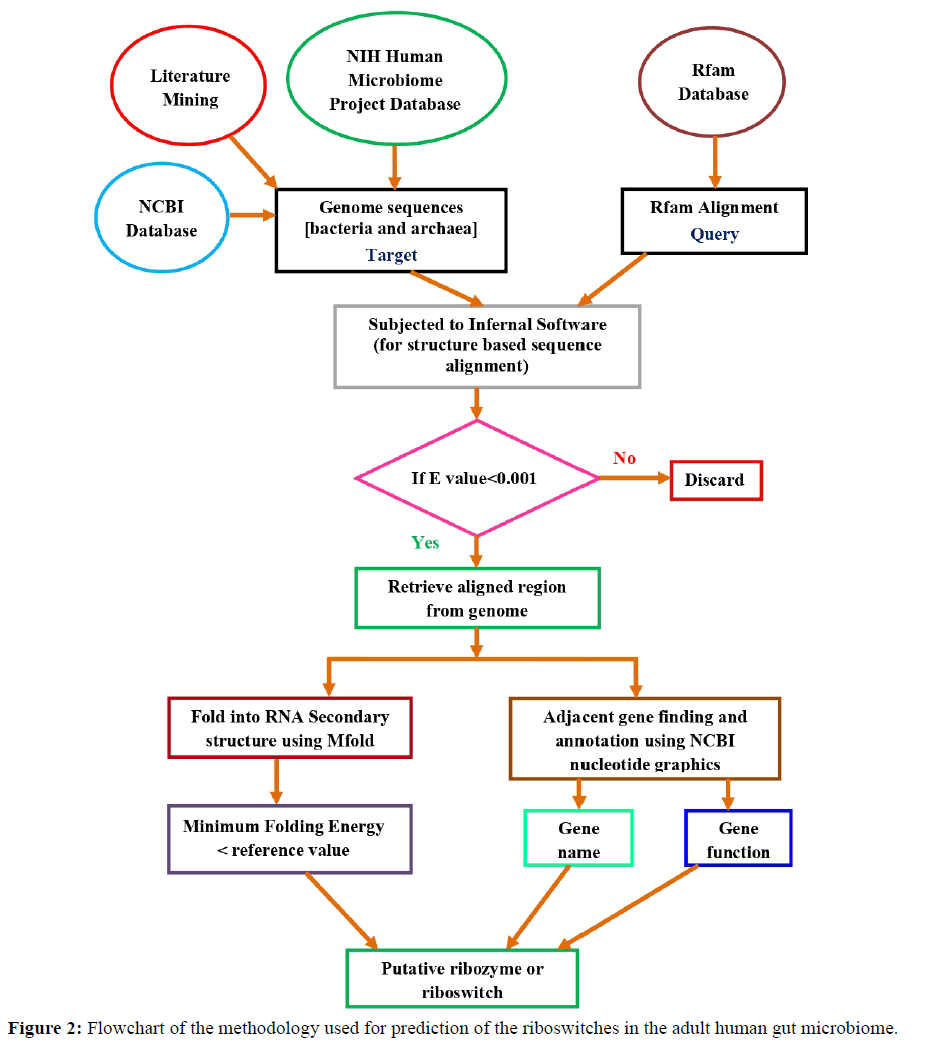
There are many tools and software available for riboswitch prediction [37]. INFERNAL software was used because it is the most reliable and accurate algorithm [38]. It is based on covariance model which is a probabilistic model that flexibly describes the secondary structure and primary sequence consensus of an RNA sequence family. INFERNAL 1.1 was used to search homologues of structural RNA sequences and to make sequence and structure-based RNA sequence alignments [39]. It forms a structurally annotated multiple sequence alignment of an RNA family with a position-specific scoring system for substitutions, insertions and deletions. The retrieved bacterial sequences were subjected to INFERNAL software to search for homologous sequences of different riboswitches. To search for closely related riboswitches, E-value was set to be less than 1e-3. E-value threshold is the expectation of random chance to yield false positive value. So, lesser E-value means less chance of getting random alignment[39, 40]. The information extracted through Infernal search are the riboswitches location in the genomes, score, E-value, GC content, and position of adjacent (upstream and downstream) genes. In this case, the score is a statistical parameter which measures sequence similarity independent of query sequence length and database size, and is normalized based on raw pairwise alignment score. The higher the score, the better is the sequence similarity [40]. The instances of the riboswitch which align to the microbial genome are referred as hits. The hit regions within the set E-value conditions were retrieved using NCBI. Then these sequences were folded using MFold, a secondary structure prediction tool for DNA and RNA based on thermodynamics [41]. The free energy of folding was noted along with the structure formed.
Simultaneously the hits were further verified for the presence of ORFs/exons 500 nucleotides downstream or upstream from the UTR region by using NCBI nucleotide graphics. The functions of the exons/genes were obtained using uniprot database [42]. A flowchart of the methodology is summarized in Figure 2.
Identification of riboswitches as potential drug target
The genomes were divided into two categories: pathogenic and non-pathogenic. Then the pathogenic microbes’ riboswitch regulated genes were studied for their role in main biosynthetic pathway, alternate pathway or as transporter proteins using BioCyc database and B. subtilis metabolic pathway reference [43]. This information along with non-pathogenic microbes’ riboswitch distribution and analogs availability provides the probable potent riboswitch as drug target. The criteria for filtering the riboswitches as suitable drug target are: they should regulate critical genes involvement in the synthesis of essential metabolites for pathogenic bacteria; should not have any alternate pathway for synthesis of essential metabolites which is not under the control of same riboswitches; should control active transport protein encoding genes of these metabolites; should be either absent or rarely present (less than 20% of the genomes) in the non-pathogenic colony of the gut; should have analog with adverse effect on gene regulation. However, the riboswitches controlling genes of active transport protein of the metabolites and having analogs are optional. If the pathogenic bacterium has some other mechanism of metabolism/synthesis of the essential metabolite or alternate pathway other than the main biosynthetic pathway then targeting that riboswitch is insignificant. If the analogs of the metabolite with adverse effect on gene regulation by riboswitch are known then it further strengthens the potential of riboswitch as drug target.
Results and Discussion
Predicted riboswitches
Screening of 63 genomes for the presence of 23 types of riboswitches resulted in identification of 545 candidate riboswitches depicted in Figure 3 showing the distribution of the riboswitches in all the genomes. The hits reported in Supplementary Table S3 showed high scores and lower e-values (E<0.001) indicating the strong possibility of the presence of particular riboswitch in the given genomic sequences. The result shows that most abundant riboswitch is TPP riboswitch (25%) followed by Cobalamin (18%), FMN (11%) and Lysine riboswitch (8%). The lower abundance is shown by YkkC/yxkD leader (2%), Cyclic di-GMP II (1%) and ZMP/ZTP riboswitch (1%). The rare ones included M. florum (0.4%), Nico (0.2%), AdoCbl variant (0.2%), and SAM-I/IV variant riboswitch (0.2%). Glutamine and preQ1-III riboswitches were not found in any of the genomes. The archaeal genomes show a few riboswitches namely fluoride, FMN and Ykok riboswitch. We further predicted the genes regulated by these riboswitches and found 494 protein coding genes, 49 hypothetical protein encoding genes and three pseudo genes (data shown in Table S3 and Supplementary File S1). The functional study of the adjacent genes was conducted which showed that they were genes encoding receptor proteins, channel proteins, adhesins, kinases, enzymes, periplasmic proteins, and flagellar/ pilin rod proteins and play role in transportation, biosynthesis and metabolic processes [44]. These analyses further resulted in the prediction of seven riboswitches as potent drug targets.
GC% analysis
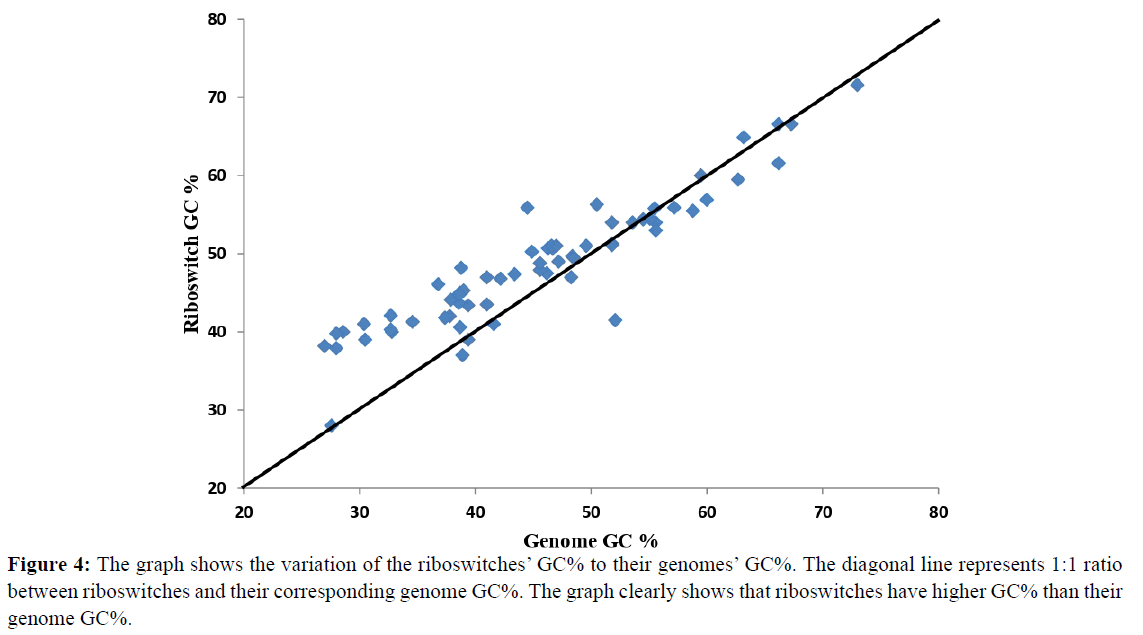
GC% of the riboswitches and their corresponding genomes was analysed by plotting a 1:1 proportional graph between riboswitch and genome GC% (Figure 4). Seventy three percent of the genomes showed greater riboswitch GC% than their corresponding genomes while 27% genomes showed lower riboswitch GC%. In our study, 40% of genomes belong to Firmicutes which are characterized by low GC content [32] therefore the graph showed high riboswitch GC% than genome GC%. Further analysis revealed that 12 microbes have considerably high riboswitch GC% than their genome GC% (7-12%). For example, in Clostridium botulinum its riboswitches have 39.8% GC content whereas its genome has 28% GC content. Similarly for Campylobacter jejuni subsp. jejuni its riboswitches GC% is 40% while the genome GC% is 30.4%.
Distribution of the riboswitches
The number of hits (riboswitch) in the genomes usually varied from 2 to 10. However in few cases such as Clostridioides difficile (46 hits), Clostridium botulinum (34 hits), Blautia coccoides (19 hits), Listeria monocytogenes (18 hits), and Lactobacillus reuteri (9 hits) showed a high number of hits that indicates high level of regulation which might be related to their pathogenicity as all the mentioned bacteria are pathogenic and found in higher numbers in the human gut in diseased condition except Lactobacillus reuteri which is used as probiotics [45].
Soutourina [46] showed expression of c-di-GMP-responsive riboswitches in C. difficile and their regulatory role in coordinated control of motility and biofilm formation. Our study also revealed presence of 12 c-di-GMP I and 4 c-di-GMP II riboswitches in C. difficile.
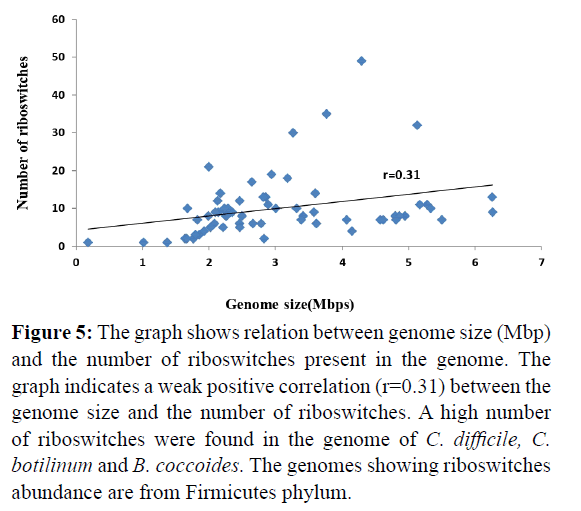
The relation between numbers of presence of riboswitches with their genome size was analysed. It was found that the number of riboswitches distribution either in different genomes or repeats in one genome is independent of the size of the genome (there is weak correlation r=0.31 between the genome size and riboswitch distribution shown in Figure 5). For example, Pseudomonas aeruginosa having 6.26 Mbp genome size shows 8 hits whereas Lactobacillus reuteri of 1.99 Mbp genome size shows 9 hits.
Figure 5: The graph shows relation between genome size (Mbp) and the number of riboswitches present in the genome. The graph indicates a weak positive correlation (r=0.31) between the genome size and the number of riboswitches. A high number of riboswitches were found in the genome of C. difficile, C. botilinum and B. coccoides. The genomes showing riboswitches abundance are from Firmicutes phylum.
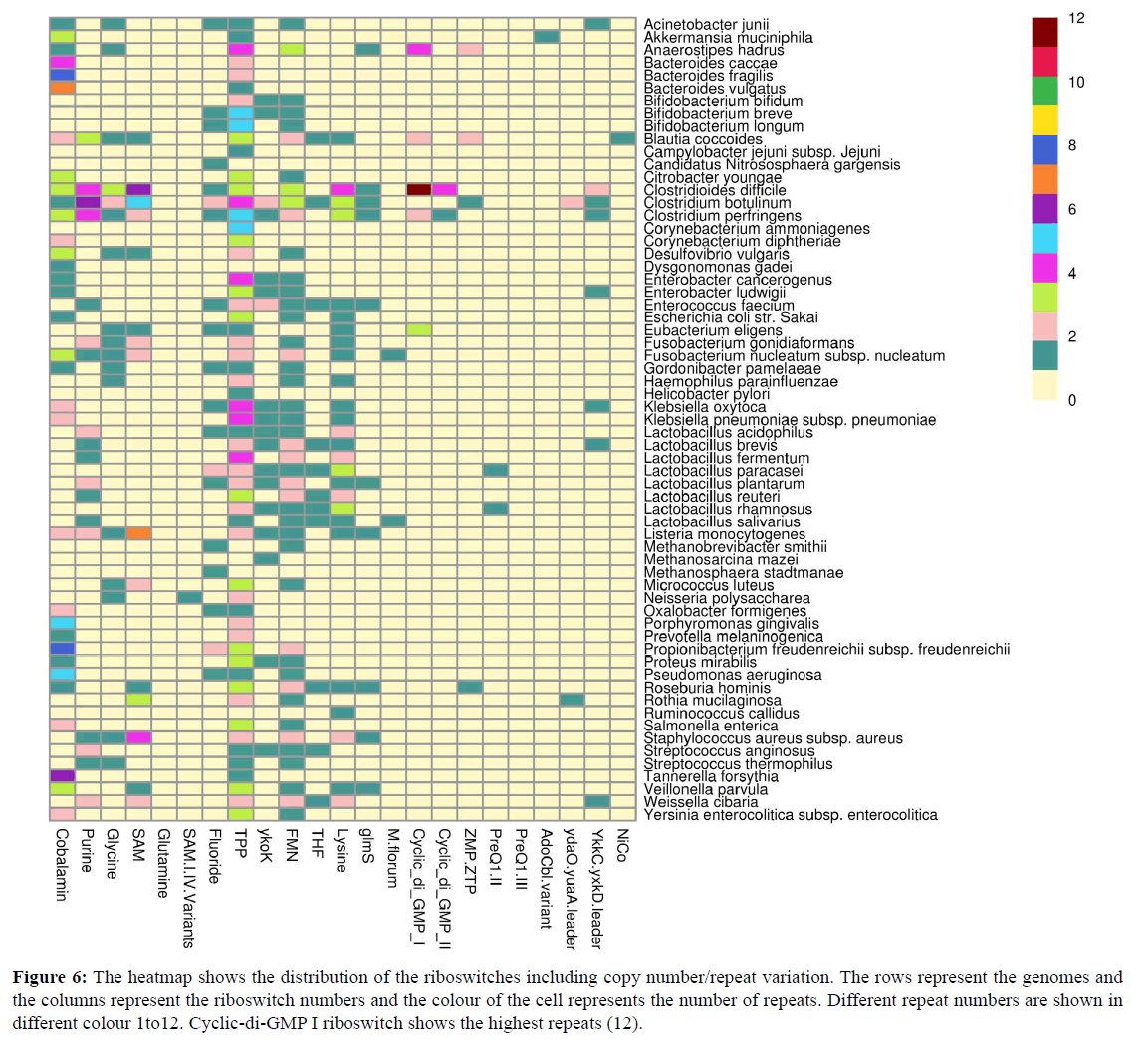
The number of instances of presence of a specific riboswitch in a single genome is shown with different colours in Figure 6. Cyclic di-GMP II showed a highest 12 times presence in C. difficile genome. More number of instances ensured different gene regulations by a particular riboswitch [47].
Figure 6: The heatmap shows the distribution of the riboswitches including copy number/repeat variation. The rows represent the genomes and the columns represent the riboswitch numbers and the colour of the cell represents the number of repeats. Different repeat numbers are shown in different colour 1to12. Cyclic-di-GMP I riboswitch shows the highest repeats (12).
Therapeutic application of the riboswitches
Finding riboswitches could be of great benefit as they play very important sensory and regulatory functions, and can be used for medicinal purposes as antibacterial drug targets for the prevention of disease because of their high specificity, wide range of target selection and action before protein translation [48]. Thus different classes of both natural and artificial riboswitches have been studied and claimed to be used as specific suppressors of gene functions and can be associated to disease-related genes (treatment of cancer, infectious diseases and genetic disorders) [49]. Recently Pavlova et al. analysed riboswitches as antibacterial drug target for antibiotic resistant human pathogens. The suitability of drug target was based on their involvement in the synthesis of essential metabolites for pathogenic bacteria, their control on active transport protein of these metabolites and on the absence of alternative metabolic pathways that are not under the control of riboswitches. Therefore, in this study antibacterial drug targets for gut pathogens had been identified on similar criteria with an additional criterion that the riboswitch acting as drug target should be present in the pathogen but it should be either absent or rarely present (less than 20% of the genomes) in the non-pathogenic colony of the gut. Seven riboswitches were identified as suitable antibacterial drug target derived from Table 1 which is discussed here.
| Riboswitch | Genes related to biosynthetic pathway | Genes related to transporter proteins | Genes related to alternative pathway | Analogs availability | Non-pathogenic microbe distribution |
|---|---|---|---|---|---|
| Cobalamin | + | + | + | Yes | 45% |
| Purine | + | - | + | - | 22.50% |
| Glycine | + | + | - | - | 20% |
| SAM | + | + | - | Yes | 17.50% |
| Fluoride/ crcB | + | + | - | - | 32.50% |
| TPP | + | + | - | Yes | 85% |
| ykoK | + | + | - | - | 25% |
| FMN | + | + | - | Yes | 65% |
| THF | + | + | - | - | 20% |
| Lysine | + | + | - | Yes | 40% |
| glmS | + | - | - | - | 10% |
| Cyclic di-GMP-I | + | + | - | - | 7.50% |
| Cyclic di-GMP-II | + | - | - | - | - |
| ydaO/yuaA leader | + | - | - | - | - |
| YkkC/yxkD leader | - | + | - | - | 10% |
Table 1: Criteria for prediction of different riboswitches as potent drug targets for pathogenic microbes of human gut. “+” sign represent the presence of the genes and –sign represent the absence. The non-pathogenic distribution less than or equal to 20 are highlighted for their significance in choosing riboswitch as drug target.
Cyclic-di-GMP riboswitches
The cyclic diguanylate (cyclic-di-GMP I) riboswitch binds to secondary messenger cyclic diguanylate and found in five genomes of which two are pathogenic namely Clostridium perfringens and Clostridioides difficile. This riboswitch controls the expression of genes involved in the metabolism of cyclic-di-GMP which play crucial role in bacterial survival. The cyclic-di-GMP I riboswitch is involved in the mechanisms such as virulence, mobility, quorum sensing and biofilm formation [50]. They are present in wide variety of bacteria specifically the pathogenic strains of proteobacteria. It also has a variant- cyclic- di-GMP II riboswitches which are present in only two genomes of bacteria-Clostridium perfringens and Clostridioides difficile. The genes regulated by them are related to proteins such as transporter binding domain, extracellular solute binding, calcium binding adhesion, flg B, Hx1R transcription regulator, zinc metallo proteases and hypothetical proteins. These riboswitches are suitable antibacterial drug target as they regulate essential transporter and metabolic genes and present only in pathogenic bacteria with 4-12 times appearance in a single genome thus regulating many genes.
GlmS riboswitch
The glucosamine-6-phosphate (glmS) riboswitch bind to cofactor glucosamine-6-phosphate is present in 10 genomes of which 7 are pathogenic. It acts as both a ribozyme and a riboswitch. The genes regulated by them are genes encoding glucosamine-fructose-6-phosphate aminotransferase and glucosamine-fructose-6-phosphate transaminase [51]. The genes are involved in the glycolysis pathway and cell wall synthesis pathway of bacteria, thus are essential genes.
SAM riboswitch
A total of 40 S-Adenosyl methionine (SAM) riboswitches were found in 15 genomes of which eight were pathogenic. SAM riboswitches bind to coenzyme s-adenosyl methionine and regulate methionine and cysteine biosynthetic pathway. The genes regulated by them include metK, metT, adenosyl methionine transferases, adenosyl methionine synthases. metK plays role in S-adenosyl-L-methionine biosynthesis.
Glycine riboswitch
The Glycine riboswitches are grouped into amino acid binding riboswitches and bind to glycine amino acid. They were distributed in 17 genomes of which 9 are pathogenic. They are usually present in the form of dimmers and regulate the genes related to symporter proteins, glycine permeases, glycine dehydrogenases and glycine degradation operon gcvT. Since glycine riboswitch presence is more in pathogenic bacteria (9 out of 23) and less in non-pathogenic microbes (8 out of 40) and regulates genes of glycine degradation operon so can act as potential drug target.
Ydao/yuaA leader riboswitch
The ydaO/yuaA leader riboswitch binds to guanidine and is present in eight genomes. Out of the eight genomes, four are pathogenic. The genes regulated are pyridine nucleotide disulphide family oxidoreductase, cell wall hydrolase and NlpC/ p60 family protein encoding gene. The cell wall hydrolase is related to the disruption of cell wall and is vital for survival thus can act as drug target. It is also not present in non-pathogenic species so further enhances their possibility as drug target.
THF riboswitch
The Tetrahydrofolate (THF) riboswitch are found in 11 genomes out of 63 genomes and are present in three pathogenic gut bacterial genomes. The genes regulated are folate ECF transporter and hypothetical protein encoding gene [52]. The transporter gene is involved in folate metabolism, thus can act as drug target.
The other riboswitches which are abundantly present in the microbes and control essential genes may also act as potential antibacterial drug target against human disease causing bacteria. So the other important riboswitches were also discussed further.
TPP riboswitch
The TPP riboswitches were most abundant (25% of all riboswitches) and found in 57 genomes. The TPP riboswitch binds to thiamine pyrophosphate coenzyme. The genes regulated by TPP mainly include thiC, thiM, thiG, and thiD which functions as thiamine phosphate synthase, energy coupled thiamine transporter protein, ABC transporter ATP binding protein, and ABC transporter permease respectively [53]. The genes belong to the TPP biosynthesis pathway and are important as drug targets.
Cobalamin riboswitch
Cobalamin riboswitch is found in second highest number (17% of all riboswitches). Cobalamin regulates the gene related to biosynthesis of vitamin B12 receptor protein where vitamin B12 is cobalamin/cynocobalamin [54]. The genes regulated by cobalamin mainly include cbiA, pduQ and cbiM which functions as cobyrinic acid a,c-diamide synthase, NADPH dependent butanol dehydrogenase, cobalamin transport protein, and receptor proteins. However cobalamin riboswitch also regulates hypothetical protein encoding genes. These genes belong to the aerobic and anaerobic cobalamin biosynthetic pathway.
FMN riboswitch
FMN riboswitch controls biosynthesis and transport proteins related to flavin mononucleotide [55]. Its presence was also reported in archaeal genome namely Methanobrevibacter smithii. Hypothetical protein genes were also regulated by FMN riboswitches. The genes include ribD, ribB and hypothetical proteins genes [56]. Some functions include riboflavin biosynthesis 5-amino-6- (5-phosphoribosylamino) uracil reductase, 3, 4-dihydroxy-2- butanone-4-phosphate synthase (DHBP synthase) functions during riboflavin biosynthesis. The gene ribD is an important component of FMN inhibition pathway of B. subtilis.
Lysine riboswitch
This class of riboswitches include amino acid binding riboswitches. The distribution pattern of lysine riboswitches showed that they are present in the 26 genomes. The genes regulated by lysine riboswitches belong to DAP biochemical pathway for synthesis of lysine, for example lysP, lysC, dapA, and asd [57]. Since the genes controlled by lysine riboswitch belong to the biosynthesis pathway the lysine riboswitch can act as potential drug target but not for human gut microbiome as this riboswitch is also widely distributed among other essential commensal non-pathogenic bacterial genomes (16 out of 40).
Purine riboswitch
Purine riboswitches were diversely distributed in 18 genomes and regulate alternative pathways. The genes regulated by purine riboswitches are purC, guaB, permease related such as xanthine /uracil permease, purine permease, aminohydrolase family permease, acetyl transferases, synthases, deaminases, and reductases which play role in cysteine and methionine metabolism pathways. Due to the presence of alternate pathways they are less suitable as drug target.
Preq1-II and preQ1-III riboswitches
These riboswitches are purine related riboswitches. In our study only preQ1-II was found in Lactobacillus paracasei and Lactobacillus rhamnosus which regulate hypothetical protein and QueT transporter family protein respectively.
Conclusion
In this study, the distribution pattern of riboswitches in the archaeal and bacterial genomes of human gut microbiome had been identified. The study revealed 545 candidate riboswitches belong to 23 different types of riboswitches present in 63 microorganisms residing in the human gut and also the adjacent genes being regulated by the riboswitches were identified and annotated. It was observed that the presence of riboswitches in a particular genome is independent of its genome size. It is found that high numbers of riboswitches were present in the pathogenic bacteria. The GC% analysis showed that the GC% of the riboswitches is higher than their genomes GC%. The following riboswitches c-di-GMP I, c-di-GMP II, glmS, SAM, THF, YdaO/YuaA leader, and glycine were identified as potent drug target in pathogenic bacteria of the adult human gut microbiome. This study can be extended in future by analyzing the expression level of these riboswitches in healthy and diseased human gut microbiome and also the finding of this work such as the distribution of riboswitches, the genes regulated by them and their functions can be archived into a database for future applications.
Acknowledgement
We thank anonymous reviewer for various useful comments. We also thank Dr. Shaoli Das and Mr. Amresh Sharma for various helpful suggestions. AS thanks DBT, India for financial support. PK thanks the UGC, India for providing financial assistant to carry out her research work.
Conflict of Interest
The authors declare no conflict of interest.
References
- Thursby E and Juge N. 2017. Introduction to the human gut microbiota. J Biochem, 474, 1823-1836.
- Cani PD. 2018. Human gut microbiome: Hopes, threats and promises. Gut, 1716-1725.
- Lewandowska M. 2010. Microbiota of human gastrointestinal tract. J Sci Tech Lodz, 74, 4-15.
- Jandhyala SM, Talukdar R, Subramanyam C, et al. 2015. Role of the normal gut microbiota. World J Gastroenterol, 21, 8836-8847.
- Eckburg PB, Bik EM, Bernstein CN, et al. 2005. Diversity of the human intestinal microbial flora. Science, 308, 1635-1638.
- Tremaroli V and Bäckhed F. 2012. Functional interactions between the gut microbiota and host metabolism. Nat, 489, 242.
- Fujimura KE, Slusher NAN, Cabana MD, et al. 2010. Role of the gut microbiota in defining human health. Expert Rev Anti Infect Ther, 8, 435-454.
- Guinane CM and Cotter PD. 2013. Role of the gut microbiota in health and chronic gastrointestinal disease: Understanding a hidden metabolic organ. Therap Adv Gastroenterol, 6, 295-308.
- Zhang YJ, Li S, Gan RY, et al. 2015. Impacts of gut bacteria on human health and diseases. Int J Mol Sci, 16, 7493-7519.
- Lozupone C, Stomabaugh J, Gordon J, et al. 2012. Diversity, stability and resilience of the human gut microbiota. Nat, 489,220-230.
- Hillman ET, Lu H, Yao T, et al. 2017. Microbial ecology along the gastrointestinal tract. Microbes Environ, 32, 300-313.
- Kho ZY and Lal SK. 2018. The human gut microbiome: A potential controller of wellness and disease. Front Microbiol, 9, 1835.
- McCown PJ, Corbino KA, Stav S, et al. 2017. Riboswitch diversity and distribution. RNA, 23, 995- 1011.
- Pavlova N, Kaloudas D, Penchovsky R, et al. 2019. Riboswitch distribution, structure, and function in bacteria. Gene, 708, 38-48.
- Waters LS and Storz G. 2011. Regulatory RNAs in bacteria. Cell, 136, 615-628.
- Guil S and Esteller M. 2015. RNA-RNA interactions in gene regulation: The coding and noncoding players. Trends Biochem Sci, 40, 248-256.
- Winkler WC, Cohen CS, Breaker RR et al. 2002. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci Am, 99, 15908- 15913.
- Barrick JE and Breaker RR. 2007. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol, 8, 11.
- Winkler WC and Breaker RR. 2003. Genetic control by metabolite-binding riboswitches. ChemBioChem, 4, 1024-1032.
- Mulhbacher J, Brouillette E, Allard M, et al. 2010. Novel riboswitch ligand analogs as selective inhibitors of guanine- related metabolic pathways. PLoS Pathog, 6, e1000865.
- Lee ER, Blount KF, Breaker RR, et al. 2009. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol, 6, 187-194.
- Ott E, Stolz J, Mack M, et al. 2009. The RFN riboswitch of bacillus subtilis is a target for the antibiotic roseoflavin produced by streptomyces davawensis. RNA Biol, 6, 276- 280.
- Serganov A, Huang L, Patel DJ, et al. 2009. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nat, 458, 233.
- Sudarsan N, Cohen CS, Nakamura S, et al. 2005. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem Biol, 12, 1325-1335.
- Blount KF, Wang JX, Lim J, et al. 2007. Antibacterial lysine analogs that target lysine riboswitches, Nat Chem Bio, 3, 44.
- Blount KF and Breaker RR. 2006. Riboswitches asantibacterialdrug targets. Biotechnology, 12, 1558.
- Mehdizadeh E, Saeid M, Barzegar A, et al. 2016. Riboswitches : From living biosensors to novel targets of antibiotics. Gene, 592, 244-259.
- Zhang L, Wong SH, Chan MTV, et al. 2017. Potential and use of bacterial small RNAs to combat drug resistance: A systematic review. Infect and Drug Resist, 521-532.
- Barh D, Tiwari S, Jain N, et al. 2011. In silico subtractive genomics for target identification in human bacterial pathogens. Drug Dev Res, 72, 162- 177.
- Hossain MU, Omar TM, Alam I, et al. 2018. Pathway based therapeutic targets identification and development of an interactive database campynibase of Campylobacter jejuni rm1221 through non- redundant protein dataset. PLoS ONE, 13, 1-23.
- Forster SC, Browne HP, Kumar N, et al. 2016. HPMCD: The database of human microbial communities from metagenomic datasets and microbial reference genomes. Nucleic Acid Res, 44, 604-609.
- Belizário JE and Napolitano M. 2015. Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches. Front Microbiol, 6, 1050.
- Shade A, Jones SE, Caporaso JG, et al. 2014. Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. Mol Bio 5, 01371-01314.
- Shetty SA, Hugenholtz F, Lahti L, et al. 2017. Intestinal microbiome landscaping: Insight in community assemblage and implications for microbial modulation strategies. FEMS Microbiol Rev, 41:182-199.
- Nawrocki EP, Burge SW, Bateman A, et al. 2014. Rfam 12.0: Updates to the RNA families database. Nucleic Acid Res, 43,130-137.
- Forster SC, Browne HP, Kumar N, et al. 2016. HPMCD: The database of human microbial communities from metagenomic datasets and microbial reference genomes. Nucleic Acid Res, 44, 604-609.
- Antunes D, Jorge NA, Caffarena ER, et al. 2018. Using RNA sequence and structure for the prediction of riboswitch aptamer: A comprehensive review of available software and tools. Front Genet, 8, 231.
- Clote P. 2015. Computational prediction of riboswitches. Methods Enzymol, 553, 287-312.
- Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics, 29, 2933-2935.
- Pearson WR. 2013. An introduction to sequencesimilarity(“homology”) searching. Curr Protoc Bioinformatics, 42, 3-11.
- Zuker M. 2003. Mfold web server for nucleic 6a1c8idfolding and hybridization prediction. Nucleic Acid Res, 31, 3406-3415.
- UniProt Consortium. 2008. The universal protein resource (UniProt). Nucleic Acid Res, 36, 190-195.
- Karp PD, Billington R, Caspi R, et al. 2017. The BioCyc collection of microbial genomes and metabolic pathways. Brief Bioinform.
- Kazanov MD, Vitreschak AG, Gelfand MS, et al. 2007. Abundance and functional diversity of riboswitches in microbial communities. BMC genomics, 8, 347.
- Markowiak P, Sliżewska K. 2017. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients, 9,1021.
- Soutourina OA, Monot M, Boudry P, et al. 2013. Genome wide identification of regulatory RNAs in the human pathogen clostridium difficile. PLoS Genet, 9.
- Pavlova, N and Penchovsky R. 2019. Genome wide bioinformatics analysis of FMN, SAM-I, glmS, TPP, lysine, purine, cobalamin, and SAH riboswitches for their applications as allosteric antibacterial drug targets in human pathogenic bacteria. Expert Opin Ther Targets, 23, 631-643.
- St pierre P, Lafontaine DA. 2010. Therapeutic applications of ribozymes and riboswitches. Curr Opin Pharmacol, 10, 551- 556.
- Abera G, Berhanu G, Tekewe A, et al. 2012. Ribozymes: Nucleic acid enzymes with potential pharmaceutical applications: A review. Pharmacophore, 3, 164-78.
- Valentini M and Filloux A. 2016. Biofilms and cyclic di-GMP (c-di-GMP) signaling: Lessons from Pseudomonas aeruginosa and other bacteria. J Biol Chem, 291, 12547-12555.
- Watson PY and Fedor MJ. 2011. The glmS riboswitch integrates signals from activating and inhibitory metabolites in vivo. NatStruct Biol, 18, 359.
- Ames TD, Rodionov DA, Weinberg Z, et al. 2010. A eubacterial riboswitch class that senses the coenzyme tetrahydrofolate. Chem Biol, 17, 681-685.
- Donovan PD. 2018. TPP riboswitch-dependent regulation of an ancient thiamin transporter in Candida. PLoS Genet, 14, e1007429.
- Vitreschak AG, Rodionov DA, Mironov AA, et al. 2003. Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA, 9, 1084-1097.
- Winkler W, Nahvi A, Breaker RR, et al. 2002. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nat, 419, 952- 956.
- Pedrolli D, Langer S, Hobl B, et al. 2015. The ribB FMN riboswitch from escherichia coli operates at the transcriptional and translational level and regulates riboflavin biosynthesis. FEBS J, 282, 3230-3242.
- Rodionov DA, Vitreschak AG, Mironov AA, et al. 2003. Regulation of lysine biosynthesis and transport genes in bacteria: Yet another RNA riboswitch? Nucleic Acid Res, 31, 6748-6757.