Review Article - Journal of Systems Biology & Proteome Research (2022) Volume 3, Issue 4
Improved interpretation of ligand occupancy and protein conformational differences
Gourab Ranjan Das*Department of Biotechnology, Utkal University, Odisha, India
- *Corresponding Author:
- Mbamalu ON
Department of Biotechnology
Utkal University, Odisha, India
E-mail: kgourab942@gmail.com
Received: 27-Jun-2022, Manuscript No. AASBPR-22-70903; Editor assigned: 30-Jun-2022, PreQC No. AASBPR-22-70903(PQ); Reviewed: 15-Jul-2022, QC No. AASBPR-22-70903; Revised: 22-Jul-2022, Manuscript No. AASBPR-22-70903(R); Published: 29-Jul-2022, DOI: 10.35841/aasbpr-3.4.117
Citation: Das GR. Improved interpretation of ligand occupancy and protein conformational differences. J Syst Bio Proteome Res. 2022;3(4):117
Abstract
Proteins and complexes that are present in natural or native-like environments can now be uniquely structurally characterised by chemical cross-linking in complex samples, cells, or even tissues. Based on sequence homology, the public database XLinkDB automatically maps crosslinks to accessible structures. Cross-linked residues must be separated by Euclidean distances within the maximal span of the applied cross-linker in order to generate the structures that are most likely to reflect protein conformations in the cross-linked sample. The solvent accessible surface distance (SASD), as opposed to the Euclidean distance, is a better predictor of consistency because it takes into account the accessibility of the cross-linked residues and the path connecting them. However, SASDs of structures are computationally expensive to calculate and not publicly available. In this section, we explain in XLinkDB version 4.0 the automatic calculation of SASDs using Jwalk for all cross-links mapped to structures, both with and without reference to ligands, and we estimate empirical maximum SASD spans for BDP-NHP and DSSO cross-linkers of, respectively, 51 and 63. We highlight cross-links sensitive to ADP binding in mitochondria isolated from HEK293 cells and show how SASDs can be utilised to help predict sample protein conformations and ligand occupancy. We also describe ligands close to cross-links in structures. A key task in medicinal chemistry and drug development comprehends the dynamics of ligands attached to proteins. However, X-ray crystallography, which is the most common method for figuring out protein-ligand structures, cannot adequately account for dynamics and cannot precisely characterise the motions of ligands in protein binding sites. In order to comprehend the conformational variability of ligands in protein crystal structures, ensemble refinement, a different technique, is employed in this work on six protein-ligand complexes.
Keywords
Chemical cross-linking, Protein structures, Solvent accessible, Surface distance, Mass spectrometry, Protein complexes, Ligands ADP, Cross-Link database.
Introduction
First, a pipeline was developed to simulate various protein topologies, both with and without ligands. These models evaluated protein structural flexibility. The models showed that the flexibility of various protein regions varies when ligands attach to proteins, and that this change occurs consistently. Conversely, proteins that becomes less rigid in other, distant locations become more stiff in the region where they interact with their ligands. In other words, ligand binding may affect how well a ligand attaches to a protein since the rest of the molecule can make up for any changes in flexibility. This research shows that ligands can modify the overall structure of the proteins they bind to, and as a result, therefore sheds new light on the role of proteins’ innate conformational flexibility during this process [1]. These findings will further our knowledge of how ligands and proteins involved in various biological processes interact with one another and, maybe, how to control these relationships. Here, using a sizable, high-quality dataset of matched holo and apo X-ray crystallographic structures, we investigate how protein sidechain conformational heterogeneity evolves upon ligand interaction. With the use of crystallographic order factors and a standardised multiconformer modelling approach, we combine both harmonic and anharmonic disorders [2].
We investigate the claim that ligand interaction reduces side chain heterogeneity in the holo structure compared to the apo structure by narrowing the conformational ensemble. Upon ligand interaction, our study uncovers intricate patterns of conformational variability that differ between and within proteins. Particularly, distant residues tend to become less rigid in proteins when binding site residues stiffen following ligand interaction. This finding implies that both natural and synthetic ligands have the ability to alter the free energy of binding by altering the natural composition of protein conformational heterogeneity throughout the whole receptor [3].
Designing novel synthetic ligands that bind to a particular protein implicated in a disease and modulate its action is a key objective of medicinal chemistry. The goal of computational drug design is to produce ligands with a strong affinity for the specified protein target. Structure-based drug design has become the norm thanks to recent technological advancements, although some aspects of it are still difficult to estimate, such as the ligand's flexibility and the entropy and free energy of binding [4]. This is due to the fact that protein ligand binding is a complicated process that is controlled by both changes in the atomic dynamics displayed by the ligand and the protein as well as particular molecular interactions between the ligand and the protein [5].
X-ray crystallography can be used to establish the atomic structures of protein-ligand complexes, which is useful for learning more about the molecular interactions between the ligand and the protein, but dynamics data is still hard to come by. All of the molecules in the crystal and the data collecting period are averaged to produce protein X-ray crystal structures, however because protein molecules are neither static nor completely organised, there is chaos. Therefore, although being complicated by various sources of disorder, the X-ray crystallographic data do contain some information about the underlying atomic dynamics. In crystallographic refinement, this information is partially considered in two different ways. The probability density distribution of an atom around its average position is first measured by atomic displacement parameters (ADPs or B factors). Additionally, alternate conformations provide discrete conformations of certain atom groupings, where each has a specific occupancy that can be optimised to a certain extent. Typically, the total number of occupancies is limited to 1.
In this paper, we use ER of X-ray crystal structures to investigate the dynamics of three combinations of ligands in the binding site of the carbohydrate-recognition domain of galectin-3 (galectin-3C). A glycan-binding protein called galectin-3 is involved in the movement of proteins, signalling, cell adhesion, angiogenesis, activation of macrophages, and apoptosis. It has been discovered to be involved with a number of illnesses, including Alzheimer's disease, inflammatory diseases, and cancer. Galectin-3C makes a good model system for crystallographic investigations because to its C-terminal domain's ability to produce massive, well-diffracting crystals in addition to its biological use. Additionally, we contrast the data on conformational variety from ER with dynamics data from other sources, including computational and experimental data from MD simulations and qFitligand. Our findings demonstrate that ER can reveal qualitative differences in ligand conformational diversity in the binding site, even when the chemical structure of the ligands differs only slightly, and may also suggest that the flexibility of some parts of the ligand and the surrounding protein is greater than suggested by standard crystallography [6].
It has been proposed that direct proof of CS can be found in the observation of the active conformation in the absence of the ligand, which is comparable to receptor constitutive activity. Direct binding to the later conformation cannot take place, at least in circumstances when the active conformation has a closed binding pocket, according to critics of CS. A partially closed conformation has occasionally been detected by sensitive probes like paramagnetic relaxation enhancement or in computer simulations of molecular dynamics [7]. The expanded CS model, which has been updated to reflect this, proposes that the protein ligand system evolves to the bound state with the closed binding pocket after the ligand binds to the partially closed conformation. Extended CS blurs the line between CS and IF, although technically speaking, the latter is an IF model since the ligand binds to an inactive conformation before the protein adopts the ultimate active conformation with the closed binding pocket. In fact, a protein with a closed binding pocket in its active conformation cannot function through a rigorous CS mechanism [8].
In any case, the active conformation in the unbound state cannot be interpreted as evidence for the CS process by simple observation. Every configuration, including the active conformation, has an equilibrium probability according to the Boltzmann distribution [9]. The amount of the equilibrium probability and the possibility of observing the active conformation as well as the sensitivity of the probe. The simple fact that the probe has improved in sensitivity should not cause the binding process to alter. Following the relative translation of the protein ligand and the internal motion of the protein from the unbound state until two reactant molecules form the bound product will unmistakably reveal the binding process and the final binding reaction [9]. Protein conformational transitions have been handled as gating as the most basic model, which means that the changes between two conformational states are roughly modelled as rate processes. Initially, it was thought that interactions between proteins and ligands had little impact on transition speeds. Recent years have seen the realisation that protein-ligand interactions always affect conformational transition rates, and that this influence is a crucial component of molecular recognition. The dual-transition-rates model Figure 1 is the consequence of various values being ascribed to the transition rates depending on whether the ligand is inside or outside the binding pocket [10].
We found top-notch, high-resolution (2 resolution or better) X-ray crystallographic datasets from the PDB to evaluate the variations in conformational heterogeneity following ligand binding. When a ligand included 10 or more heavy atoms, we classed a structure as holo, eliminating frequent crystallographic additions Figure 1. Structures without ligands, excluding common crystallographic additives, were classified as apo Figure 1.
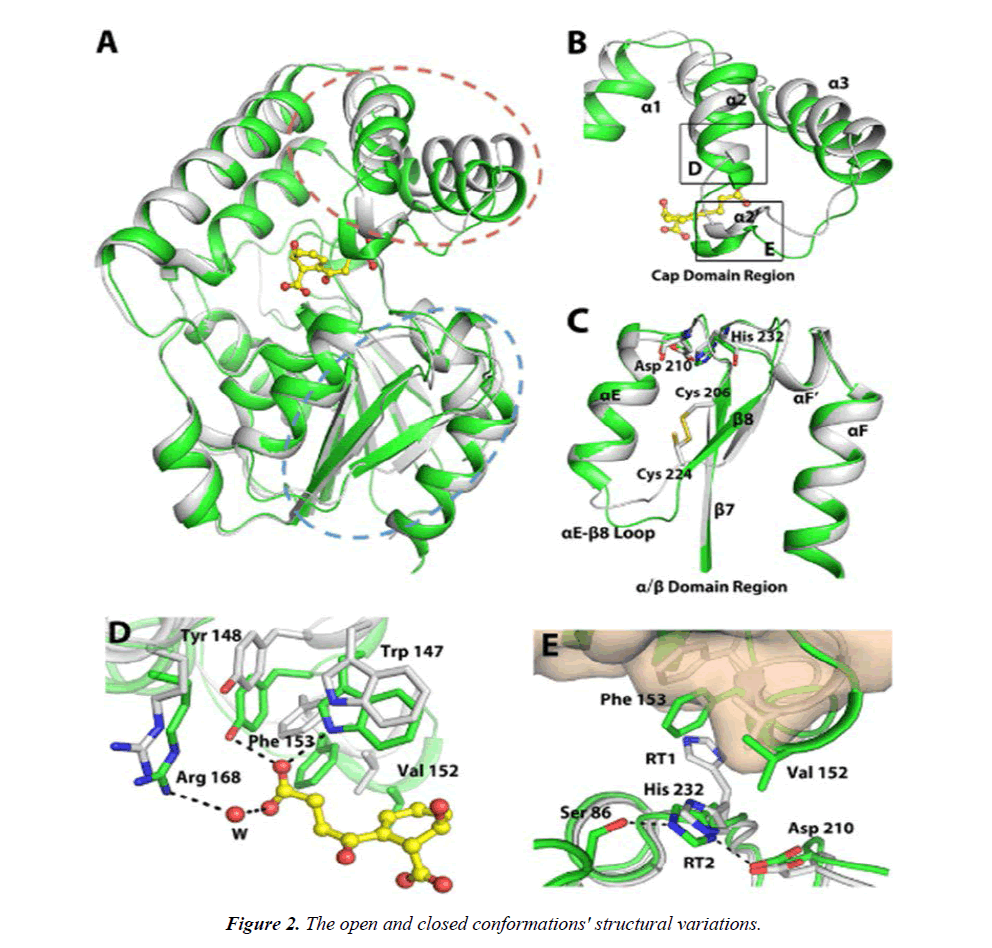
A, combining the two structures. B, the ensemble in the cap domain that contains the numbers 2 and 3. C, backbone movement on a grand scale in the /-domain. D, the hinged movement allows for interactions between enzymes and SHCHC. E, the 2 3 10-helix's interaction with the triad residues. Figure 2 shows the closed MenH-SHCHC structure in green for all C atoms and the open ligand-free MenH structure in grey. The active site residues are shown as sticks in Figure 2. The areas circled in broken red and blue lines in A, respectively, correspond to B and C. The RT1 rotamer of His 232 is omitted in C. D and E correspond to the boxed regions in B. In E, the -helical cap domain of the open conformation is presented in wheat surface. Dashed lines indicate hydrogen bonds with a distance less than 3.5 Å. Water molecules (W) are shown as red spheres.
In our investigation of 13 inhibitor-bound datasets of the kinase CDK2, in the same crystal form and space group, we found a complicated interplay between conformational change and dynamics. The value of crystal systems susceptible to isomorphous soaking or co-crystallization is highlighted by the possibility to investigate a single protein with several ligands. Between type I and type II inhibitors, we found variances in conformational heterogeneity that may be categorised with well-known alterations, such as variations in the DFG motif. The pattern of protein-protein interactions on distal surfaces, which is crucial for the development of CDK inhibitors, is affected by the tuning of distal site dynamics, which may be a viable technique for modifying the affinity of kinase inhibitors [11].
In terms of drug design, ER supports the widely held belief that maintaining the native ligands' galactose moiety is crucial for maintaining their affinity for galectin-3C, but changing the glucose moiety in synthesised ligands can improve their ability to interact with other protein residues [12]. We also produced alternative conformations for the lactose ligand using the qFitligand software, which uses a conformational search within the ligand's degrees of freedom to generate new conformations that fit the experimental electron density, to ensure that the numerous conformations for the lactose ligand's glucose moiety in the ER simulations are not an artefact [13,14].
Electrostatic forces are not taken into account, and statistical geometry restrictions rather than an energy-based force field are employed for the bonds, angles, and dihedrals during ER's MD simulations [15]. We also ran MD simulations of the lactose-galectin-3C complex to determine whether this unconventional force field has an impact on the ER outcomes. The MD trajectory of lactose-galectin-3C exhibits significant flexibility, with movements occurring mostly in the lactose glucose moiety. Although several extreme conformations of the glucose component are absent in the MD simulations, this is comparable to what was seen in the ER [16]. This could be because the length of the conventional MD simulations prevents them from overcoming the obstacles required to obtain these conformations or because they are artefacts brought about by the unconventional force field employed in the ER simulations. However, solution MD simulations also occasionally overestimate the dynamics of the ligand in comparison to ER because the simulations are carried out in solvent rather than the crystal (so that symmetryrelated molecules are missing) and there are no restrictions on the movement of the atoms, in contrast to ER, where the movements are constrained by the crystallographic data. For instance, the protein's flexibility is greater [17].
Conclusion
As a result, our findings suggest that ER provides information that is complimentary to that provided by conventional crystallographic refinement. It highlights the areas of the structure that are ill-defined and could experience significant variations. It provides a clear picture of the conformational diversity and the various potential conformations. Such details are crucial for understanding the structure and point out areas where the one-conformation perspective might be incorrect. Additionally, ER identifies the protein-ligand interactions that are negligible and weak for binding. This is crucial for medication development since it is ideal to use the strongest interactions in the binding site when developing new synthetic medicines.
References
- Caldararu O, Kumar R, Oksanen E, et al. Are crystallographic B-factors suitable for calculating protein conformational entropy?. Physic Chem Chemi Phys. 2019;21(33):18149-60.
- Zavodszky MI, Lei M, Thorpe MF, et al. Modeling correlated main‐chain motions in proteins for flexible molecular recognition. Struct Funct Genet and Bioinf. 2004;57(2):243-61.
- Srinivasan J, Miller J, Kollman PA, et al. Continuum solvent studies of the stability of RNA hairpin loops and helices. J Biomol Struct. 1998;16(3):671-82.
- Sherman W, Day T, Jacobson MP, et al. Novel procedure for modeling ligand/receptor induced fit effects. J of Medici Chemi. 2006;49(2):534-53.
- Englebienne P, Moitessier N. Docking ligands into flexible and solvated macromolecules. 4. Are popular scoring functions accurate for this class of proteins?. J Chemi Inform and Mode. 2009;49(6):1568-80.
- Murray CW, Baxter CA, Frenkel AD. The sensitivity of the results of molecular docking to induced fit effects: application to thrombin, thermolysin and neuraminidase. J of Comp Aided Mole Desi. 1999;13(6):547-62.
- Hoffmann D, Kramer B, Washio T, et al. Two-stage method for protein− ligand docking. J Medic Chem. 1999;42(21):4422-33.
- Bowman AL, Lerner MG, Carlson HA. Protein flexibility and species specificity in structure-based drug discovery: dihydrofolate reductase as a test system. J the Ameri Chemi Soci. 2007;129(12):3634-40.
- Case DA, Cheatham III TE, Darden T, et al. The Amber biomolecular simulation programs. J Computa Chem. 2005;26(16):1668-88.
- Eisenhaber F, Argos P. Improved strategy in analytic surface calculation for molecular systems: Handling of singularities and computational efficiency. J Computa Chem. 1993;14(11):1272-80.
- Ferrara P, Gohlke H, Price DJ, et al. Assessing scoring functions for protein− ligand interactions. J Medic Chem. 2004;47(12):3032-47.
- Bahar I, Lezon TR, Yang LW, et al. Global dynamics of proteins: bridging between structure and function. Annual Rev of Biophy. 2010;39:23.
- Keating KS, Flores SC, Gerstein MB, et al. StoneHinge: hinge prediction by network analysis of individual protein structures. Prote Sci. 2009;18(2):359-71.
- Rader AJ, Hespenheide BM, Kuhn LA, et al. Protein unfolding: rigidity lost. Proceedings of the National Academy of Sciences. 2002;99(6):3540-5.
- Dunitz JD. Win some, lose some: enthalpy-entropy compensation in weak intermolecular interactions. Chem & Biol. 1995;2(11):709-12.
- Huggins DJ. Quantifying the entropy of binding for water molecules in protein cavities by computing correlations. Biophys J. 2015;108(4):928-36.
- Chang CE, Chen W, Gilson MK. Evaluating the accuracy of the quasiharmonic approximation. J Chemi The and Comput. 2005;1(5):1017-28.
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref