Mini Review - Journal of Biochemistry and Biotechnology (2021) Volume 4, Issue 1
Importance of detection systems of real-time PCR for novel coronavirus-19 diagnosis.
Jai Prakash Muyal*, Basav Hazarika
School of Biotechnology, Gautam Buddha University (GBU), Greater Noida, Uttar Pradesh, India
- Corresponding Author:
- Jai Prakash Muyal
School of Biotechnology
Gautam Buddha University (GBU)
Greater Noida, Uttar Pradesh, India
Tel: +911202344273
E-mail: jmuyal@gbu.ac.in
Accepted date: December 22, 2020
Citation: Muyal JP, Hazarika B. Importance of detection systems of real-time PCR for novel coronavirus-19 diagnosis. J Biochem Biotech. 2021;4(1):1-5
Abstract
The novel coronavirus (COVID-19) is a contiguous infectious disease, caused by SARS-CoV2. The presence of this new virus was unknown before the outbreak began in Wuhan, a major metropolitan city of China, in December 2019. As per WHO, it has been recently characterized as a pandemic disease, and would be a one of the agents of potentially deadly diseases that are a great public health threat of 21st century. As the occurrence of COVID-19 is quite new, hence the appropriate and early detection of this virus is a major challenge for the doctors and scientists alike. However, so far there are several detection tools for the covid-19 infection which are being in use includes methods that detect the presence of the virus itself (RT-PCR and antigen detection) and those that detect antibodies produced in response to infection. Nevertheless all these methods are having some pros and cons. Therefore, a reliable, fast and low cost diagnosis tool is urgently required for the detection of COVID-19 infection. In this review article, we emphasize the significance of real-time PCR technology, which is one of the finest options for the detection of presence of viral genome in patients. However, real-time PCR further relies on different detection chemistries that play a critical role in screening out a reliable result in cost effective manner.
Keywords
COVID-19, Real-time PCR, Detection systems of real-time PCR.
Introduction
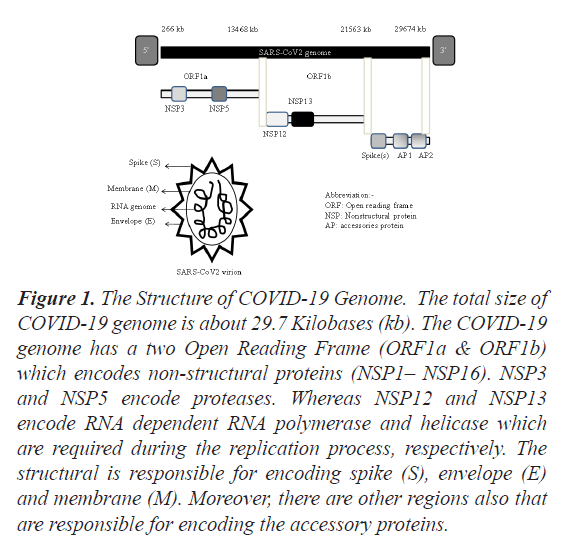
Coronaviruses are a large family of viruses having RNA as a genome and may disease in mammals and birds. So far only seven coronaviruses are known to cause disease in humans. Out of these seven coronaviruses, three of them {(Middle East Respiratory Syndrome (MERS), Severe Acute Respiratory Syndrome (SARS) and Coronavirus disease (COVID-19)} can be causes mild or brutal respiratory tract infections and gastrointestinal diseases, and have caused major outbreaks of deadly pneumonia in the 21st century. As per the World Health Organization (WHO) Regional Office for Europe, on 7th January, 2020, a novel coronavirus (nCoV) was first identified and was temporarily named ‘2019-nCoV’. Later on, it was named the ‘Covid-19 Virus’. The genome size of COVID-19 is between 26 to 32 kilobases [1]. The 5′end of the genome is having a methylated cap while a 3′end of the genome is having a polyadenylated tail. Nevertheless, the detailed structure of COVID-19 genome organization has been shown in Figure 1.
Figure 1: The Structure of COVID-19 Genome. The total size of COVID-19 genome is about 29.7 Kilobases (kb). The COVID-19 genome has a two Open Reading Frame (ORF1a & ORF1b) which encodes non-structural proteins (NSP1– NSP16). NSP3 and NSP5 encode proteases. Whereas NSP12 and NSP13 encode RNA dependent RNA polymerase and helicase which are required during the replication process, respectively. The structural is responsible for encoding spike (S), envelope (E) and membrane (M). Moreover, there are other regions also that are responsible for encoding the accessory proteins.
The World Health Organization declared the outbreak of coronavirus-19 (Covid-19) as a pandemic in March, 2020. It is a novel respiratory infection with serious clinical manifestations, including death. The COVID-19 epidemic came in the month of December, 2019 in Wuhan City in Hubei territory of the country [2]. There are evidences which prove that the original spread of COVID-19 infection from bats to humans started towards the end of 2019. Consequently the disease spread to more states in China, and thereby to the rest of the world. As per the WHO, COVID-19 Dashboard, as of 3rd December 2020, there have been 65.4 million confirmed cases of COVID-19, including 1.51 million deaths, worldwide.
According to current evidence, COVID-19 affects different people in different ways. The COVID-19 is principally transmitted between one to another person through respiratory droplets and contact routes and thus, it is highly contagious [3]. Most of the infected people develop only mild to moderate illness and can even recover without hospitalization. According to the Centres for Disease Control and Prevention (CDC), COVID-19 infection leads to certain symptoms which usually appear about 2 to 14 days after the first exposure to the virus. They includes fever, cough, difficulty in breathing, repeated shaking with chills, muscle pain, headache, sore throat, loss of taste or smell, skin rashes, bluish lips. However, existence of these symptoms does not necessarily indicate an infection. Unfortunately, till now, there is no specific vaccine or drugs are available against COVID-19 infection. Only socially distancing is the only way to prohibit virus transmission from the person-to-person.
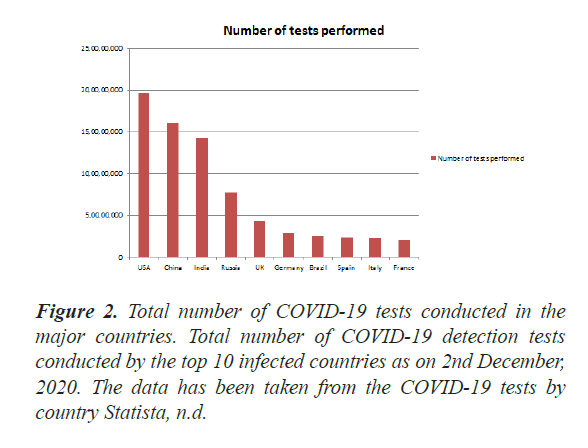
Early detection of the COVID-19 in the patient is one of the challenges towards its eradication and safe life. There are several diagnosis tools for the COVID-19 infection which are being in use for such purposes, however all those tools are having some merits and demerits. Therefore, a reliable and less cost effective tool is required for COVID-19 detection, so that a maximum population can be screened out worldwide. Today diagnostic testing for COVID-19 is becoming increasingly available through commercial and hospital-based laboratories in addition to public health laboratories. So far, about more than 30 million diagnostic tests for COVID-19 detection were conducted worldwide (Figure 2).
Two different methods are now approved globally for the presence of COVID-19 detection in a test sample. The first method directly detects the presence of the SARS CoV-2 virus itself while in the second method detects antibodies which are produced by the immune cells when the body is exposed to an infection. However, an antibody test is not as effective because, firstly, it takes around 6-21 days for the body to make antibodies after an infection, secondly doesn’t provide a precise idea of an infection types. Beside that some immunoassays method have also been suggested for SARS CoV-2 virus antigens or antibodies detection. However they also failed to maintain the specificity, reliability and reproducibility [4]. Not only this, a next-generation sequencing approach has also been tried out towards the detection of COVID-19 [5]. However, this technology is so expansive, several steps in between need to be optimized and required skilled personnel. Since several factors are being associates with these technologies, and therefore an urgent requirement of a suitable alternative method is needed for effectively detection of the presence of the virus in the test sample.
Alternatively, Polymerase Chain Reaction (PCR) based viral RNA detection approach is considered to be one of the best molecular diagnostic tools for COVID-19 detection due to its rapid, easy operational, highly sensitive characteristics [6]. Therefore, PCR-based molecular detection of a virus is currently the most preferred method by healthcare personnel as well as researchers. However the real-time reverse transcription-polymerase chain reaction (Real-time PCR) is the ‘gold standard’ frontline test for COVID-19 due to relatively simple and tremendous specific in nature [7] than the conventional PCR technology. This technology combines the PCR principle with the use of fluorescent reporter molecules in order to scrutinize the production of amplification products during each cycle of the PCR reaction. Thus, the combination of exceptional sensitivity, reproducibility, and low contamination risk has made real-time PCR technology a tempting tool for the COVID-19 detection. The detection systems which are available in real-time PCR further add quality and costing concerns. Thus in this review, we have tried to compare two different types of detection systems of real-time PCR method towards screening of large number of population for the COVID-19 detection. We believe this comparison will certainly help researchers/technicians in dissecting the cost and reliability in selection of the detection system associated with this method for mankind.
Real-time PCR technology using SYBR Green I chemistry
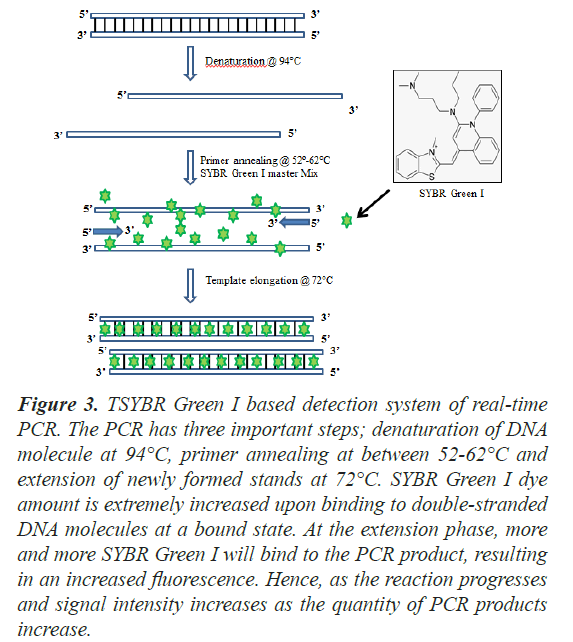
Mostly SYBR Green I (or rarely Eva Green) is a commonly used fluorescent dye that binds double-stranded DNA molecules by intercalating between the DNA bases, thus widely used in real-time PCR technology. It is used in Real-time PCR because the emitted fluorescence can be measured at the end of each amplification cycle to determine the relatively amount of copy number of test gene. It is a cyanine dye and is asymmetrical in nature and is also structurally related to other dsDNA-specific dyes such as YOYO-1 and TOTO-1 [8]. The binding affinity of SYBR Green I to nucleic acid is about 100 times more than that of ethidium bromide dye. It has been reported that when SYBR Green I intercalates with double stranded DNA, the fluorescence of SYBR Green increases manifold. SYBR Green I exclusively binds sequence to the minor groove of double stranded DNA. It has an excitation wavelength of around 480 nm and a maximum emission wavelength of around 520 nm followed by a quantum yield of 0.8 [9]. The fluorescence of this bound dye is at least 1000 times higher than that of the free dye. Hence, it is highly useful to monitor the accumulation of PCR product [10]. Upon monitoring in real time, an increase in the fluorescence signal can be observed during the polymerization step which slowly decreases along with the denaturation of DNA (Figure 3). The amount of DNA product keeps on increasing along with the amplification reactions and so does the number of SYBR green molecules. The increase in fluorescence is directly proportional to the amount of PCR product. Moreover, the measurement of fluorescence needs to be done at the end of the elongation step of each PCR cycle. This approach gets rid of the use of targetspecific fluorescent probes. Therefore, this method can be employed with any pair of primers for any target and hence making it cost-effective [11].
Figure 3: TSYBR Green I based detection system of real-time PCR. The PCR has three important steps; denaturation of DNA molecule at 94°C, primer annealing at between 52-62°C and extension of newly formed stands at 72°C. SYBR Green I dye amount is extremely increased upon binding to double-stranded DNA molecules at a bound state. At the extension phase, more and more SYBR Green I will bind to the PCR product, resulting in an increased fluorescence. Hence, as the reaction progresses and signal intensity increases as the quantity of PCR products increase.
Advantages and disadvantages of using SYBR Green in real-time PCR
SYBR Green I dye has several advantages over conventional dyes used in real-time PCR. Some of these dyes include greater sensitivity and ease of use as gels soaked in diluted stain can be visualized without desalting. It is also non-carcinogenic in nature and hence, relatively safer to use. It is also cost-effective that is much cheaper and easier to use than TaqMan probe. However, a major disadvantage of SYBR Green I is that its specificity is determined only by the primers. As a result, one has to consider the risk of amplifying non-specific PCR products during optimization [12]. It may generate non-specific reaction product, which might further yield false positive results. This also requires the generation of a melting curve by plotting fluorescence as a function of temperature in order to verify the final PCR product [13]. Further downstream processing of PCR is involved as it requires the generation of a melting curve. Also, this system has a limited application, which includes quantification of gene expression and detection of pathogens.
Real-time PCR using TaqMan probes
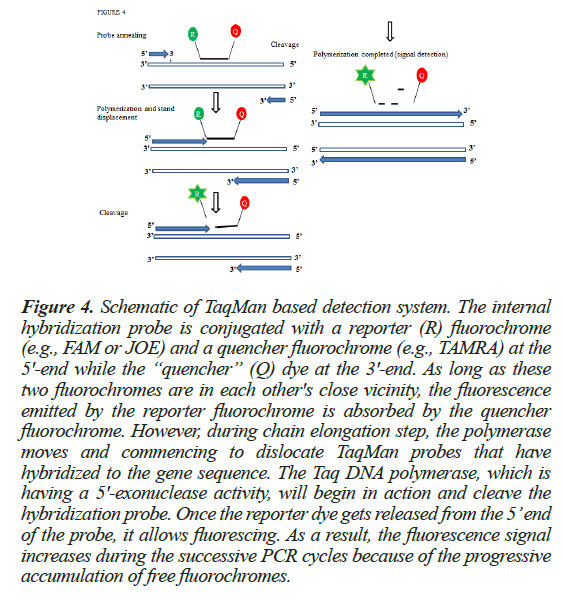
Another method to associate the PCR product with the fluorescent signal is the use of fluorescent reporter probes. The probe is nothing but an additional primer which binds specifically to the target DNA during the annealing step [14] along with the gene specific primer set. These probe-based technologies usually make use of a TaqMan probe system. TaqMan probes are hydrolysis probes that are used to increase the specificity of real-time PCR. This system is consisting of an oligonucleotide probe of a length of about 20 bp, having a reporter fluorophore at the 5’end and a quencher fluorophore at the 3’ end. Among the fluorophores, 6-carboxyfluorescein (FAM) or Tetrachlorofluorescein (TET) are available while tetramethylrhodamine (TAMRA) is generally used as the quencher [15]. When the fluorophore emits fluorescence, it is quenched by the quencher via Forster Resonance Energy Transfer (FRET). The detailed methodology has been discussed in Figure 4.
Figure 4: Schematic of TaqMan based detection system. The internal hybridization probe is conjugated with a reporter (R) fluorochrome (e.g., FAM or JOE) and a quencher fluorochrome (e.g., TAMRA) at the 5'-end while the “quencher” (Q) dye at the 3'-end. As long as these two fluorochromes are in each other's close vicinity, the fluorescence emitted by the reporter fluorochrome is absorbed by the quencher fluorochrome. However, during chain elongation step, the polymerase moves and commencing to dislocate TaqMan probes that have hybridized to the gene sequence. The Taq DNA polymerase, which is having a 5'-exonuclease activity, will begin in action and cleave the hybridization probe. Once the reporter dye gets released from the 5’ end of the probe, it allows fluorescing. As a result, the fluorescence signal increases during the successive PCR cycles because of the progressive accumulation of free fluorochromes.
In this method, the TaqMan probe principle relies on the 5´–3´ exonuclease activity of Taq polymerase. Taq DNA polymerase synthesizes new strands using the unlabeled primers and the template. During the ongoing reaction, when the DNA polymerase reaches a TaqMan probe, its endogenous 5’ nuclease activity causes cleavage of the probe and ultimately, separates the dye from the quencher. More and more dye molecules are released with each passing cycle of PCR. Finally, this leads to a substantial increase in fluorescence intensity which is proportional to the amount of amplicon. The positive results from a realtime TaqMan PCR can be visualized by two ways. The first one is the amplification plot that tells how much of the reporter dye is generated during amplification. It is directly related to the formation of final PCR product. The second one is via determining the cycle threshold (CT value), which is chiefly the intersection between the amplification plot and the threshold values. This CT value provides an amount of the target gene’s copy number present in the PCR reaction [16] of the test sample.
Advantages and disadvantages of using TaqMan probe in real-time PCR
Probably the biggest advantage of TaqMan PCR is the use of internal probes to quantify PCR products and thus detect only specific amplification products. As the hybridization step is carried out during amplification itself, there is no such requirement of post-amplification handling and thus, the risks of manual handling and carry-over are minimized [16]. Moreover, It has a very high specificity and has an absolute sensitivity of 5-10 molecules [17]. Its applications include quantification of gene expression and profiling, chromatin immunoprecipitation, Single Nucleotide Polymorphism genotyping, detection of gene mutations and detection of genome instabilities etc. However, TaqMan probe also has a disadvantage in that a different probe has to be synthesized for each unique target sequence. It is costlier as it involves the use of expensive probes.
Conclusion
COVID-19 infection is rapidly spreading across the globe, so far, infected about 2.0 million people in several countries and still spreading like anything. Since the virus made its appearance all of a sudden, several countries did not have the necessary infrastructure to detect the presence of this virus amongst individuals, an immediate requirement of reliable detection tool is required for its manifestation, timely. Even though the immunoassay detection test is easier and cheaper to use, they are not very precise and reliable. Other techniques like Next Generation Sequencing are so expansive and time consuming tools. Hence, for large scale testing of samples, real-time PCR based diagnosis has been turned out to be the most widely accepted techniques for the COVID-19 detection globally. Out of the two most common detection systems that is SYBR Green I and TaqMan probe, it has been concluded that both the detection systems has a capability to provide a clear cut signature of presence of COVID-19 viral infection in patients. However, TaqMan based real-time PCR allows COVID-19 detection in more specific manner. Nevertheless, SYBR Green I based realtime PCR detection is also fair if the cost of COVID-19 diagnosis is also a concern for large scale testing of samples. We believe in that detection of COVID-19 infected individuals preferably by real-time PCR will be continuing in near future in managing this pandemic.
Conflict of Interest
The authors declare they have no conflict of interest directly or indirectly related to the manuscript contents.
Acknowledgements
None.
References
- Woo PCY, Huang Y, Lau SKP, et al. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2(8):1804-20.
- Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3):2000045.
- Liu J, Liao X, Qian S, et al. Community Transmission of Severe Acute Respiratory Syndrome Coronavirus 2, Shenzhen, China, 2020. Emerg Infect Dis. 2020;26(6).
- Tang YW, Schmitz JE, Persing DH, et al. The Laboratory Diagnosis of COVID-19 Infection: Current Issues and Challenges. J Clin Microbiol. 2020;58(6).
- Chen L, Liu W, Zhang Q, et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg Microbes Infect. 2020;9(1),313-9.
- Tahamtan A, Ardebili A. Real-time RT-PCR in COVID-19 detection: Issues affecting the results. Expert Review of Molecular Diagnostics. 2020;453-4.
- Shen M, Zhou Y, Ye J, et al. Recent advances and perspectives of nucleic acid detection for coronavirus. Journal of Pharmaceutical Analysis. 2020.
- Jin X, Yue S, Wells KS, et al. Sybr Green (Tm)-1-a new fluorescent dye optimized for detection of picogram amounts of DNA in gels. Biophysical Journal 1994;66(2):159.
- Rodríguez-Lázaro D, Hernández. Real-time PCR in food science: Introduction. Curr Issues Mol Biol. 2013;15:25-38.
- Morrison TB, Weis JJ, Wittwer CT. Quantification of lowcopy transcripts by continuous SYBR® green I monitoring during amplification. Biotechniques. 1998;24(6):954-8.
- Giulietti A, Overbergh L, Valckx D, et al. An overview of real-time quantitative PCR: Applications to quantify cytokine gene expression. Methods. 2001;25(4):386-401.
- Simpson DAC, Feeney S, Boyle C, et al. Retinal VEGF mRNA measured by SYBR Green I fluorescence: A versatile approach to quantitative PCR. Mol Vis. 2000;6:178-83.
- Ririe KM, Rasmussen RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997;245(2):154-60.
- Kesmen Z, Gulluce A, Sahin F, et al. Identification of meat species by TaqMan-based real-time PCR assay. Meat Sci. 2009;82(4):444-9.
- Kutyavin I V. 3’-Minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 2000.
- Leutenegger CM. The Real-Time TaqMan PCR and Applications in Veterinary Medicine. Veterinary sciences tomorrow. 2009;2001.
- Hofmann-Lehmann R, Swenerton RK, Liska V, et al. Sensitive and robust one-tube real-time reverse transcriptase-polymerase chain reaction to quantify SIV RNA load: Comparison of one- versus two-enzyme systems. AIDS Res Hum Retroviruses. 2000;16(13):1247-57.