Research Article - Journal of Biochemistry and Biotechnology (2019) Volume 2, Issue 1
Immobilization of firefly luciferase on the cell plasma membrane as a quantitative biosensor for measurement of ATP in the pericellular space in live mammalian cells
Khue Vu Nguyen1,2*, Kefeng Li1, Robert K Naviaux1,2,31Department of Medicine, Biochemical Genetics and Metabolism, The Mitochondrial and Metabolic Disease Center, School of Medicine, University of California, San Diego, USA
2Department of Pediatrics, School of Medicine, University of California, San Diego, USA
3Department of Pathology, School of Medicine, University of California, San Diego, USA
- *Corresponding Author:
- Dr Khue Vu Nguyen
Department of Medicine, Biochemical Genetics and Metabolism
The Mitochondrial and Metabolic Disease Center
School of Medicine,University of California
Building CTF, Room C-103, 214 Dickinson Street
San Diego, CA92103-8467, USA
Tel: +1 619 543 2105
E-mail: kvn006@ucsd.edu
Accepted Date: February 20, 2019
Citation: Khue Vu Nguyen, Kefeng Li, Robert K Naviaux. Immobilization of firefly luciferase on the cell plasma membrane as a quantitative biosensor for measurement of ATP in the pericellular space in live mammalian cells. J Biochem Biotech 2019;2(1):1-10.
Abstract
The purinergic signaling system consists of transporters, enzymes and receptors responsible for the synthesis, release, action and extracellular inactivation of adenosine-5’-triphosphate (ATP) and its extracellular breakdown product adenosine. Up to date, the full appreciation of the role of ATP as an extracellular signal has been hampered by lack of proper biosensors for accurate local real-time measurement of extracellular ATP concentration in the pericellular space from individual cells under physiological and pathological conditions. We describe herein the development of simple, sensitive, and reliable dual-function biosensor for the local real-time measurement of extracellular ATP concentration in the pericellular space in live HEK 293 cells by performing an immobilization of firefly luciferase (Fluc) coupled with the green fluorescent protein (GFP) on the plasma membrane of HEK 293 cells via a glycosyl-phosphatidylinositol, GPI, anchor derived from the human folate receptor 1 (FOLR1) protein: pmeLUC. Our pmeLUC2 dual-function reporter construct was shown to be fluorescent and bioluminescent and could detect pericellular ATP at concentrations under physiological conditions: <5 μM and the apparent KM of immobilized Fluc for ATP are to be 2-3 × 10-6 M, values much lower than the 51 × 10-6 M found for the free Fluc. In addition, there was no loss of immobilized Fluc activity in pmeLUC2 after more than 15 ATP measurements followed by 90 days stored at +4°C in PBS. The method used for the construction of our pmeLUC2 probe may pave the way for new strategies applicable to rational pmeLUC design. Its use in live cells and organisms, especially for identifying a new pathway for ATP secretion as a signaling molecule, promise to further expand its utility.
Keywords
Firefly luciferase, Human folate receptor 1 protein, Green fluorescence protein, Glycosylphosphatidylinositol, Luminescence, Fluorescence, HEK 293 cells, ATP.
Introduction
For many years, adenosine 5’- triphosphate (ATP) was solely considered for its role as the main source of energy in living cells, we now know that ATP also plays a fundamental physiological role as a pleiotropic extracellular messenger of cell-to cell communication acting at plasma membrane receptors named purinergic receptors. The purinergic signaling system employs extracellular purines (most notably ATP, and adenosine) and pyrimidines as signaling molecules. Both purine and pyrimidine nucleotides are released from living cells via several physiologically relevant mechanisms, which include exsocytosis, diffusion through membrane channels and via transporters [1-4]. Furthermore, purines and pyrimidines are released from stressed and dying cells, being early and universal indicators of cell damage [3,5]. Today, the role of ATP as a signaling molecule is not limited to the nervous system as indeed ATP sensitivity and ATP-mediated signaling has been identified in virtually all tissues and cell types [6]. Therefore, ATP appears to be the most widespread and omnipresent of all known extracellular signaling molecules, which appeared very early in evolution [6]. Immediately after release, ATP has a half-life measured in seconds as a result of a complex array of potent ectonucleotidases and other hydrolytic activities, which degrade ATP and generate ADP, AMP and adenosine [7,8]. Once outside the cell, ATP mediates its diverse effects by binding to and activating a broad range of receptors. The actions of ATP are mediated by ionotropic P2X and metabotropic P2Y receptor subfamilies, whilst the actions of adenosine are mediated by P1 adenosine receptors [3,5]. Under basal conditions, ATP is present intracellularly in concentrations of 3-5mM. Typically, the concentration of ATP required for half-maximal activation of purinergic receptors is 3-500nM, values 1,000-fold lower than those inside the cell. Consequently, ATP released in quantities sufficient to initiate signalling does not appear to alter intracellular energy stores [9]. The essence of the purinergic signalling hypothesis is that cellular stimulation releases ATP and subsequently activates nucleotide receptors in the cell (autocrine activation) and/or in adjacent cells (paracrine activation), thereby regulating or modulating cellular functions. The actions of ATP itself, however, likely are limited to a narrow paracrine radius of a few hundred microns due to the rapid kinetics of these reactions and its dispersion by regional blood or fluid flow. Up-to-date, the full appreciation of the role of ATP as an extracellular signal has been hampered by lack of proper probes for accurate local real-time measurement of the extracellular ATP concentration close to the surface of the plasma membrane i.e. exactly in the vicinity of the cell surface (pericellular space) where extracellular ATP is biologically active from individual cells under physiological and pathological conditions.
In an attempt to provide a simple and reliable dual-function biosensor for the local real-time measurement of extracellular ATP concentration in the pericellular space in live mammalian cells, we have performed an immobilization of firefly luciferase (Fluc) coupled with the green fluorescent protein (GFP) on the cell plasma membrane via a glycosylphosphatidylinositol, GPI, anchor derived from the human folate receptor 1 (FOLR1) protein.
Material and Methods
Isolation of genomic DNA, amplification and cloning
The RNA-free genomic DNA from normal subject was isolated from the human whole peripheral blood using the Puregene® DNA purification kit (Gentra System, Minneapolis, Minnesota, USA). The DNA concentration was determined by using the ND-1000 spectrophotometer NanoDrop® device (Thermo Scientific). The genomic DNA so obtained was used for amplification of the N-terminal leader sequence (LS) and the C-terminal GPI anchor of the human FOLR1 gene using the polymerase chain reaction (PCR). The entire coding sequence (CDS) of the firefly luciferase from Photinus pyralis (Fluc, EC 1.13.12.7) was PCR amplified from the plasmid pGL4.20 (luc2/Puro) (Promega Corporation, Madison, WI 53711-5399, USA). The CDS of the green fluorescent protein (GFP) from copepod Potellina sp. was PCR amplified from the plasmid pmaxGFP (Lonza Walkerville Inc., Walkerville, MD 21793, USA). The sequences of primers used for PCR are available upon request. All obtained purified DNA fragments were first subjected to the ligation reaction into the pCR® II plasmid vector and then introduced in One Shot® TOP10 chemical competent E. coli strain for cloning by using the reagents and the transformation procedure of the TA cloning kit (Invitrogen, Carlsbad, CA, USA). The resulting vectors were termed (1), (2), (3), (4), (5), and (6) for pCR® II/LS, pCR®II/GPI (with the TGA stop codon), pCR® II/Fluc (with the TAA stop codon), pCR® II/Fluc (without the TAA stop codon), pCR® II/GFP (with the TGA stop codon), pCR® II/GFP (without the TGA stop codon) respectively.
Construction of the immobilized firefly luciferase on the cell plasma membrane (pmeLUC)
The mammalian expression vector pcDNA3.1 (+) (Invitrogen, Carlsbad, CA, USA) was used for the construction of the immobilized firefly luciferase on the cell plasma membrane (pmeLUC). All primers used for the construction of the pmeLUC are available upon request. All resulting vectors were obtained after cloning in One Shot®TOP10 chemical competent E. coli strain by using the reagents and the transformation procedure of the TA cloning kit (Invitrogen, Carlsbad, CA, USA). We first performed a PCR using (2) as template and primers allowing the creation of the XbaI and ApaI sites. The obtained PCR product was then inserted in the right frame into the pcDNA3.1 (+) vector predigested with XbaI and ApaI enzymes. The resulting vector was termed (7). The vector (4) was subjected to the digestion with BamHI and XhoI enzymes and the resulting DNA fragment was then inserted in the right frame into the vector (7) predigested with BamHI and XhoI enzymes. The resulting vector was termed pmeLUC1. Also by using the vector (6) as template, a PCR using primers allowing the creation of the NheI and HindIII sites was performed. The obtained PCR product was then inserted in the right frame into the pmeLUC1 vector predigested with NheI and HindIII enzymes. The resulting vector was termed pmeLUC2. We also performed a PCR using (1) as template and primers allowing the creation of the Acc65I and KPnI sites. The obtained PCR product was then inserted in the right frame into the pmeLUC1 vector predigested with Acc65I and KpnI enzymes. The resulting vectors were termed pmeLUC3 (with LS 1X), pmeLUC4 (with LS 2X), and pmeLUC5 (with LS 3X). Furthermore, the vectors (3) and (5) were subjected to the digestion with BamHI and XhoI enzymes and the resulting DNA fragments were then inserted in the right frame into the pcDNA3.1 (+) vector predigested with BamHI and XhoI enzymes. The resulting vectors were termed (8) and (9). These vectors (8) and (9) were used as positive controls of luciferase (free Fluc) and GFP (free GFP) activities respectively. As negative control of luciferase activity, a CDS of a non-luciferase gene containing a TGA stop codon of the p16/CDKN2A gene, inserted in the right frame into the pcDNA3.1 (+) vector between the BamHI and XhoI sites was obtained as a kind gift from Dr. Mitchell B. Diccianni, Department of Pediatrics, University of California, San Diego, USA. This vector was termed (10). In sum, by using the mammalian expression vector pcDNA3.1 (+) as backbone, the following constructs were performed: GPI anchor of FOLR1- Fluc: pmeLUC1; GPI anchor of FOLR1-Fluc-GFP: pmeLUC2; GPI anchor of FOLR1-Fluc-LS1x: pmeLUC3; GPI anchor of FOLR1-Fluc-LS2x: pmeLUC4; GPI anchor of FOLR1-Fluc- LS3x: pmeLUC5; pcDNA3.1 (+)-Fluc: 8 (positive control of luciferase activity, free Fluc); pcDNA3.1 (+)-GFP: 9 (positive control of GFP activity, free GFP); pcDNA3.1 (+)-non-Fluc gene: 10 (negative control of luciferase activity, non-Fluc).
Cell transfection
The human embryonic kidney (HEK 293) cells (ATCC, Manassas, Virginia, 20110-2209 USA) were cultured in 96 well tissue culture plate (Corning® CellBIND®, Corning, NY 14831, USA) using DMEM with 4.5 g/L glucose, L-glutamine & sodium pyruvate complemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin (all from Gibco®by Life TechnologiesTM, USA). HEK 293 cells were transfected with 100 ng DNA per well via the lipofection reagent (TransITR-293 reagent, Mirus, USA). The reagents of this kit and the reaction conditions used are according to the manufacturer’s recommendations. For cotransfection, a ratio of 1:1 was performed in which 100 ng DNA each per well was used. We also generated clones stably expressing pmeLUC by culture of the transfected cells in the presence of G418 sulfate (Geniticin®, Gibco® by Life TechnologiesTM, USA) (0.8 mg/ml added 48 hrs after transfection) for 3 weeks. Stable pmeLUC expressing clones were kept in the continuous presence of 0.4 mg/ml G418 sulphate.
Confocal fluorescence microscopy analysis and luciferase assay
36 h after transfection, HEK 293 cells were subjected to the confocal fluorescence microscopy analysis and luciferase assay. The confocal fluorescence microscopy analysis was performed using the Leica TCS SP5 confocal system (Leica Microsystems, Inc., Buffalo Grove, IL 60089, USA). Luciferase assay for checking the luciferase activity was carried out on cell lysate using the luciferase assay system kit (Promega Corporation, Madison, WI, USA). The reagents of this kit and the reaction conditions used are according to the manufacturer’s recommendations. Briefly, 100 μl of the cultured medium were first aspirated from the cells, and the well was rinsed with 200 μl of phosphate-buffered saline (PBS). After aspiration of PBS, the cells were then lysed with 20 μl of the lysis reagent at room temperature for 5 min, and 100 μl of the luciferase assay reagent were then added. The bioluminescence produced instantly was detected by using the POLARstar Omega luminometer (Polarstar Omega, BMG Labtech, Inc., NC 27513, USA). All luminescence (Units) obtained at the emission wavelength of 560 nm for measuring of luciferase activity was a mean of four determinations.
ATP measurement
Live HEK 293 clones: ATP measurement in the pericellular space in live mammalian cells was carried out using live HEK 293 clones stably expressing pmeLUC1 or pmeLUC2 cultured in 96 well tissue culture plate (Corning® CellBIND®, Corning, NY 14831, USA) and the ATP determination kit (Molecular Probes, Inc., OR 97402, USA). The reagents of this kit and the reaction conditions used are according to the manufacturer’s recommendations. For experiments, 1μl of Dluciferin and 1μl of dithiothreitol (DTT) were added into the 100 μl of cultured medium to the final concentrations of 500 μM and 1mM respectively. Then, 1 μl of different amounts of ATP was also added into the cultured medium to the final concentrations ranging from 0 μM to 200 μM ATP. The bioluminescence produced instantly was detected by using the POLARstar Omega luminometer (Polarstar Omega, BMG Labtech, Inc., NC 27513, USA). All luminescence (Units) obtained at the emission wavelength of 560 nm for measuring of luciferase activity was a mean of four determinations.
“Ghost” of HEK 293 clones: For checking the localization of the biosensors on the outer surface of the plasma membrane, ATP measurement was also carried out using the HEK 293 “ghost” (dead cell in which the outline remain visible, but without the cytosol as well as other cytoplasmic structures or stainable nucleus), and the reagents of the luciferase assay system kit (Promega Corporation, Madison, WI, USA). From the HEK 293 clones stably expressing pmeLUC1 or pmeLUC2 cultured in 100 mm X 20 mm dish (Celltreat® scientific products, China), 12 ml of the cultured medium were aspirated from the cells, and the dish was rinsed with 15 ml of PBS. After aspiration of PBS, the cells were then lysed with 1 ml of the lysis reagent at room temperature for 5 min. The HEK 293 “ghost” was obtained by scraping attached cells from the dish, transferred to a 15 ml centrifuge tube, and followed by 5 successive rinses with 1 ml of PBS, in which the “ghost” was pellet by brief centrifugation after each rinse. No luciferase activity was found from the PBS of these successive rinses. For experiments, the obtained HEK 293 “ghost” was first transferred to one well of the 96 well tissue culture plate (Corning® CellBIND®, Corning, NY 14831, USA). 100 μl of the luciferase assay reagent were then added. Then, 1μl of different amounts of ATP were also added to the final concentrations ranging from 0 μM to 200 μM ATP. The bioluminescence produced instantly was detected by using the POLARstar Omega luminometer (Polarstar Omega, BMG Labtech, Inc., NC 27513, USA). All luminescence (Units) obtained at the emission wavelength of 560 nm for measuring of luciferase activity was a mean of four determinations.
Between two ATP concentrations used, the HEK 293 “ghost” was washed by 5 successive rinses with 200 μl of PBS. After aspiration of PBS from the last rinse, 100 μl of fresh luciferase reagent were then added for ATP measurement. Otherwise, unused HEK 293 “ghost” so obtained can be stored at +40C in 1 ml of PBS.
Results
Structure and expression of Fluc from pmeLUC transfected cells
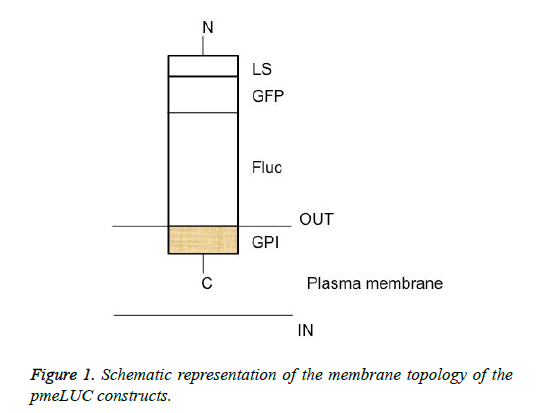
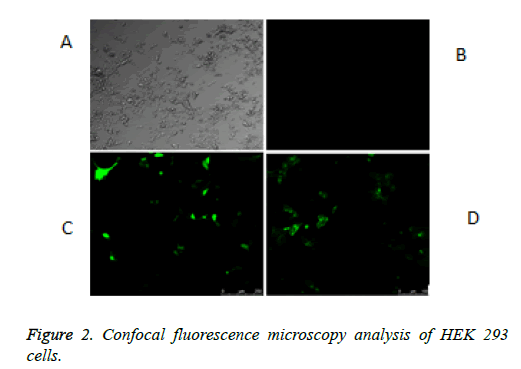
The schematic structure of the pmeLUC constructs used as biosensors for the local real-time measurement of ATP concentration in the pericellular space in live HEK 293 cells is shown in Figure 1. The confocal fluorescence microscopy analysis of HEK 293 cells non-transfected or transfected with the vector (9) or pmeLUC1, and pmeLuc2 was performed. There was no fluorescence emission from the HEK 293 cells with pmeLUC1 (Figures 2A and 2B). The presence of GFP expression seen only on the surface of (clusters) (Figure 2), compared with that seen throughout transfected cells (Figure 2C), suggests that the Fluc is expressed only on the plasma membrane [10]. This would be therefore the case for pmeLUC1, pmeLUC3, pmeLUC4, and pmeLUC5 transfected cells.
The construct comprising the C-terminal GPI anchor of the FOLR1 protein-Fluc: pmeLUC1; the C-terminal GPI anchor of the FOLR1 protein-Fluc-GFP: pmeLUC2; the C-terminal GPI anchor of the FOLR1 protein-Fluc-the N-terminal LS of the FOLR1 protein, 1X : pmeLUC3; the C-terminal GPI anchor of the FOLR1 protein-Fluc-the N-terminal LS of the FOLR1 protein, 2X : pmeLuc4; and the C-terminal GPI anchor of the FOLR1 protein-Fluc-the N-terminal LS of the FOLR1 protein, 3X : pmeLUC5.
A: HEK 293 cells. B: no fluorescence emission from the HEK 293 non-transfected or transfected with pmeLUC10: pcDNA3.1 (+)-non-Fluc gene (negative control of luciferase activity, non-Fluc i.e. unmodified expression vector). C: fluorescence emission throughout the HEK 293 cells transfected with pcDNA3.1 (+)-GFP: 9 (positive control of GFP activity, free GFP). D: fluorescence emission as many dots from the HEK 293 cells transfected with GPI anchor of FOLR1-Fluc-GFP: pmeLUC2.
Luciferase assay
Luciferase assays for checking the luciferase activity were carried out on cell lysate of HEK 293 cells non-transfected or transfected with the vectors (8), (9), (10), pmeLUC1, pmeLuc2, pmeLUC3, pmeLUC4, pmeLUC5, and cotransfected with pmeLUC1 and (9). The results obtained are shown in Table 1. There were no luciferase activities obtained with HEK 293 cells non-transfected or transfected with the vectors (9), or (10) (Table 1). By comparison to the luciferase activity obtained with (8) transfected cells, there were decreases of luciferase activities of 36, 55, 93, 98, 98% for pmeLUC1, pmeLUC2, pmeLUC3, pmeLUC4, pmeLUC5 transfected cells respectively (Table 1). The limitation in conformation changes of the immobilized Fluc via GPI anchor on the plasma membrane of pmeLUC1 transfected cells was responsible for the reduction of luciferase activity (36%). Concerning the pmeLUC2, pmeLUC3, pmeLUC4, and pmeLUC5 transfected cells, there was an addition of limitation in conformation changes of the immobilized Fluc caused by steric hindrance due to the presence of GFP in pmeLUC2, and LS in pmeLUC3, pmeLUC4, pmeLUC5, and led to a great decrease in luciferase activity: 55% due to GFP and 93-98% due to LS. Affectation by steric hindrance due to the presence of LS was then the most (93-98%), especially with LS 2X in pmeLUC4 and LS 3X in pmeLUC5: 98% (Table 1). In any case, however, we cannot rule out the possibility of lower Fluc protein expression level from these biosensors. By comparison to the luciferase activity obtained with pmeLUC1 transfected cells, a dramatic decrease in luciferase activity (69%) was observed from the pmeLUC1 and (9) co-transfected cells (Table 1). Here, a competition for entry into the cells in favor of the vector (9) was responsible for this decrease in luciferase activity. For quantitative purpose of ATP measurement in the pericellular space in live mammalian cells, it is therefore preferable to perform the transfection with a single construct containing both reporter genes: Fluc for ATP measurement and GFP for the control of transfection process. The pmeLUC2 appears then suitable for such a purpose. Taking into account for these results, the pmeLUC1 and pmeLUC2 were selected for further work in ATP measurement.
| HEK 293 cells | Luminescencea (Units) | Decreaseb (%) |
|---|---|---|
| non-transfected | 61 | - |
| transfected with (8) | 653291 | - |
| transfected with (9) | 214 | - |
| transfected with (10) | 788 | - |
| transfected with pmeLUC1 | 415208 | 36 |
| transfected with pmeLUC2 | 295139 | 55 |
| transfected with pmeLUC3 | 47476 | 93 |
| transfected with pmeLUC4 | 9689 | 98 |
| transfected with pmeLUC5 | 10131 | 98 |
| co-transfected with pmeLUC1 and (9) | 128650 | 69 |
(8): pcDNA3.1 (+)-Fluc (positive control of luciferase activity,
free Fluc i.e. no immobilization of Fluc on the membrane).
(9): pcDNA3.1 (+)-GFP (positive control of GFP activity, free
GFP i.e. no immobilization of GFP on the membrane).
(10): pcDNA3.1 (+)-non-Fluc gene (negative control of
luciferase activity, non-Fluc i.e. unmodified expression vector).
pmeLUC1: pcDNA3.1 (+)-GPI anchor of FOLR1-Fluc.
pmeLUC2: pcDNA3.1 (+)-GPI anchor of FOLR1-Fluc-GFP.
pmeLUC3: pcDNA3.1 (+)-GPI anchor of FOLR1-Fluc-LS1X.
pmeLUC4: pcDNA3.1 (+)-GPI anchor of FOLR1-Fluc-LS2X.
pmeLUC5: pcDNA3.1 (+)-GPI anchor of FOLR1-Fluc-LS3X.
aLuminescence (Units) obtained is a mean of four
determinations.
bDecreases in luciferase activities (%) for pmeLUC1,
pmeLUC2, pmeLUC3, pmeLUC4, pmeLUC5 transfeccted
cells; and for pmeLUC1 and (9) co-transfected cells are
respectively calculated by comparison to luminescence (Units)
obtained with (8) and pmeLUC1.
Table 1: Mesasurement of luciferase activity on cell lysate of HEK 293 cells.
ATP measurement
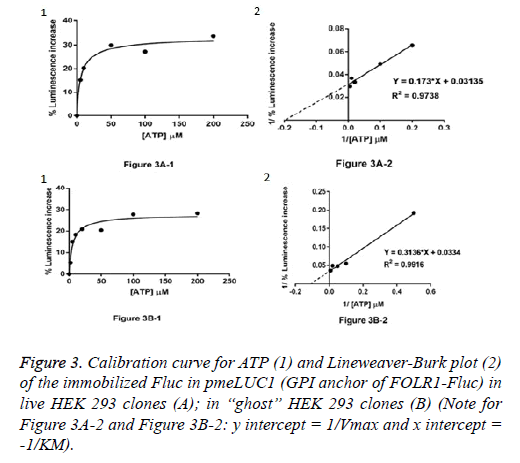
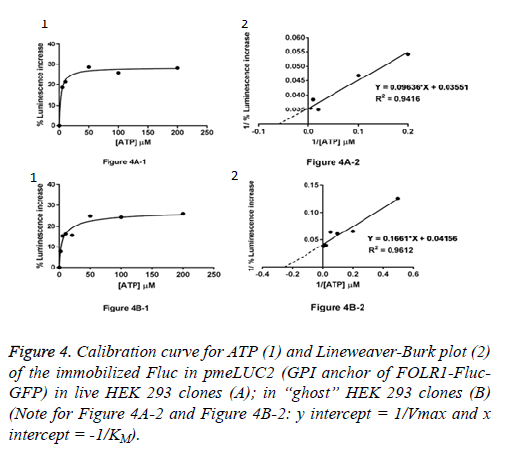
Live HEK 293 clones: ATP measurement in the pericellular space in live HEK 293 clones stably expressing pmeLUC1 or pmeLUC2 cultured in 96 well tissue culture plate was performed. Cells expressing pmeLUC have a basal level of luminescence emission that depends on the amount of expressed luciferase. After additions of D-luciferin and DTT into the cultured medium, light emission recorded corresponds therefore to ATP measurement in the pericellular space in live HEK 293 clones. Subsequent ATP additions evoke further increases in light emission that allow building a calibration curve. To minimize variations due to minor changes in pmeLUC expression, luminescence can be expressed as Figure 3A-1 for pmeLUC1 and Figure 4A-1 for pmeLUC2.
“Ghost” of HEK 293 clones: In order to checking the localization of the biosensors on the outer surface of the plasma membrane, ATP measurement was also carried out using the HEK 293 “ghost” (dead cell in which the outline remain visible, but without the cytosol as well as other cytoplasmic structures or stainable nucleus obtained after the cell lysis) of the HEK293 clones stably expressing pmeLUC1 and pmeLUC2. In the same manner, by using the HEK 293 “ghost”, the calibration curves for ATP were also obtained: Figure 3B-1 for pmeLUC1 and Figure 4B-1 for pmeLUC2. These experimental results confirm the localization of the biosensors on the outer surface of the cell plasma membrane of the pmeLUC1 and pmeLUC2 transfected HEK293 cells. Enzymatic activity detected from the biosensors pmeLUC1 and pmeLUC2 was then from the immobilized Fluc on the cell plasma membrane. This is therefore the case for pmeLUC3, pmeLUC4, and pmeLUC5 transfected cells. Indeed, the cell lysis is a method in which the outer boundary or cell membrane is broken down in order to release inter-cellular materials such as DNA, RNA, protein or organelles from a cell and it was also used by other authors for discriminating between cell membrane and cytosolic proteins [11].
Catalytic parameters: apparent KM and Vmax: From the calibration curves for ATP (Michaelis-Menten curves shown in (Figure 3A-1 and Figure 3B-1, Figure 4A-1 and Figure 4B-1), the apparent KM and Vmax values for ATP of the immobilized Fluc in pmeLUC1 and pmeLUC2 from live HEK 293 clones as well as HEK 293 “ghost” were determined graphically from Lineweaver-Burk plots (see Figure 3A-2 and Figure 3B-2, Figure 4A-2 and Figure 4B-2), and given in Table 2. Their apparent KM is similar and much lower than that of the free Fluc enzyme (3-9 x 10-6 M instead of 51 x 10-6 M) [12,13]. It is also interesting to note that the apparent KM values for ATP of immobilized Fluc on nylon tubes and epoxy methacrylate beads were 22 x 10-6 M and 6.6 x 10-6 M respectively [12,13].
| Transfected HEK 293 clones | apparent KM | apparent Vmax |
|---|---|---|
| with pmeLUC1 in live HEK 293 clones: | 5.51 Ã 10-6M | 31.89 Ã 10-6M s-1 |
| with pmeLUC1 in “ghost’’ of HEK 293 clones: | 9.38 Ã 10-6M | 29.94 Ã 10-6M s-1 |
| with pmeLUC2 in live HEK 293 clones: | 2.71 Ã 10-6M | 2.71 Ã 10-6M s-1 |
| with pmeLUC2 in “ghost” of HEK 293 clones: | 3.99 Ã 10-6M | 24.06 Ã 10-6M s-1 |
pmeLUC1: pcDNA3.1 (+)-GPI anchor of FOLR1-Fluc.
pmeLUC2: pcDNA3.1 (+)-GPI anchor of FOLR1-Fluc-GFP.
aapparent KM and Vmax values were determined from the Lineweaver-Burk plots see (Figure 3A-2 and Figue 3B-2, Figure 4A-2 and Figure 4B-2).
Table 2: Catalytic properties (apparent KM and Vmax valuesa) for ATP of immobilized Fluc on the plasma membrane of HEK 293 clones.
It is important to note herein that one of the most interesting characteristics of immobilized enzyme, compared to the free soluble form of the enzyme, was it lets enzyme be held in place throughout the reaction, following which it is easily separated from the products and may be used again as well as its increased resistance to changes in conditions such as pH or temperature and its long storage period [14,15]. In the present study, the effectiveness (allow reuse of enzyme) and stability (no loss of enzymatic activity following a long storage period) of the pmeLUC1 and pmeLUC2 biosensors on the outer surface of the HEK293 cell membrane were also verified: effective after more than 15 ATP measurements followed by 90 days stored at 40C in PBS in which the similar values of luminescence emission were found for the measurement of different ATP concentrations such as 5 μM, 10 μM, and 50 μM.
Discussion
Reporter genes, genes that encode proteins whose presence is readily detected and quantified, have significantly advanced a number of efforts in biology and biotechnology, including studies of gene regulation, gene delivery, and signal transduction [16]. The reporter genes used initially, such as chloramphenicol acetyltransferase and galactosidase, has gradually yielded to more sensitive, non-radioactive reporters based on fluorescence and luminescence. These include fluorescent proteins, such as the green fluorescent protein (GFP) from the jelllyfish Aequorea victoria, and luciferases, including firefly luciferase (Fluc) from Photinus pyralis [17,18].
GFP has the advantages that its intrinsic fluorescence is readily visualized, and that it is non-enzymatic and thus does not require a substrate; however, considerable concentrations of GFP (≈ 1 μM) must be present inside the cell to detect a signal over the background noise [19,20]. Furthermore, it is highly stable intracellularly, with a half-life of over 24 hours. However, its stability poses a significant disadvantage for dynamic studies of short time scale gene expression events, and GFP variants with lower half-lives have a correspondingly lower sensitivity [21]. Fluc catalyzes the reaction of Dluciferin with O2 to produce light in the presence of Mg2+ and ATP. Fluc is used to measure ATP and to control ATPproducing and consuming systems [22]. Luciferases have the advantage of a very low background noise, thus decreasing the number of molecules needed for a detectable signal. This attribute is particularly useful in studies of promoters with low or transient activity. The relatively short half-life reported for luciferase can also serves a practical purpose when the enzyme is utilized to study the dynamics of gene expression [23]. Because of these advantageous properties, luciferases have been employed in a wide variety of studies including gene delivery [24], growth factor regulation of gene expression [25], and in live cells and organisms [26,27]. Some available commercial kits to quantifying ATP such as Agilent Seahorse XFP Real-Time ATP Rate Assay, Kit No 103591-100; Cayman Chemical ATP Detection Assay Kit-Luminescence, Kit No 700410; Abcam ATP Assay Kit (Colorimetric/Fluorometric), Kit No ab83355; Abcam Luminescent ATP Detection Assay Kit, Kit No 113849; Molecular Probes® ATP Determination Kit (Invitrogen), Kit No A22066 as well as the use of HPLC method [28,29] or HPLC method coupled with fluorescence detection [30]. However, all these methods were designed to measure total ATP levels in living cells and did not allow realtime measurement of extracellular ATP concentration in the pericellular space. Measurement of the extracellular ATP concentration has rapidly become a frequent application of the standard luciferine/luciferase assay, given the increasing importance that purinergic signaling has recently achieved in cell biology [3,5,9]. However, the measurement of the extracellular ATP concentration with soluble luciferase has two major limitations: in the first place, sample manipulation causes a perturbation that by itself might cause unwanted cell stimulation with consequent release of ATP; second, soluble luciferase is likely to be unable to detect rapid changes in the concentration of ATP in the pericellular space. Thus, there is a need to develop novel probes/techniques that allow closer monitoring of ATP kinetics in the pericellular space under physiological and pathological conditions. In 2005, Pellegatti, P. et al. [31] performed the construction of pmeLUC for extracellular ATP measurement in live HEK 293 cells in which the CDS of Fluc (from Photinus pyralis) was inserted in-frame (between GPI anchor of FOLR1 and myc tag) into the preexisting sequence of the GPI anchor of FOLR1-myc tag-LS of FOLR1. The whole sequence was cloned into the mammalian expression vector pcDNA3: pcDNA3-GPI anchor of FOLR1-Fluc-myc tag-LS of FOLR1. This pmeLUC biosensor corresponds to the pmeLUC3 of our present work. However, the detail regarding the experimental information to obtaining the LS and GPI anchor of FOLR1 as well as myc tag sequences was not available. The resulting pmeLUC probe so obtained had a low affinity for ATP determination: allowed measurement only above the 5-10 μM ATP level i.e. above the pericellular ATP concentration under physiological conditions [31] and could not detect ATP in the pericellular space from healthy tissues [32]. In the present work, our pmeLUC3 probe: pcDNA3.1 (+)-GPI anchor of FOLR1-Fluc-LS1X showed 93% reduction in luciferase activity (see Table 1). This finding demonstrated that the presence of LS in the pmeLUC probe obtained by Pellegatti et al. [31] created therefore drawbacks in luciferase activity (allowed measurement only above the 5-10 μM ATP level [31]) as well as affinity (apparent KM=47 x 10-6M), calculated from the figure 7B in [32]) resulted in high detection limit for ATP and could not consequently detect significant extracellular ATP levels in healthy tissues [32]. These authors started their work with the whole final construct of pmeLUC3 without studying the effects of each of the components of the construct separately for measurement of ATP. They could not see therefore these drawbacks of the pmeLUC3 (biosensor with LS). In fact, the problems of LS and GPI are complex. Proteins are produced on ribosomes and can be divided into two general groups based upon whether or not LS (sometimes referred to as signal peptide or leader peptide) is encoded at their amino terminus. Proteins without LS are translated on “free” ribosomes and may remain in the cytosol. Proteins specifying an N-terminal LS complete translation on endoplasmic reticulum (ER)-attached ribosomes (or “bound” ribosomes) and will either stay in the ER, the Golgi, vacuoles, or be secreted to the plasma membrane, cell wall or extracellular matrix. The LS is a short peptide (usually 16-30 amino acids long) [33] present at the N-terminus of the majority of newly synthesized proteins that are destined towards the secretory pathway [34]. Although most type I membrane-bound proteins have LS, the majority of type II and multi-spanning membrane-bound proteins are targeted to the secretory pathway by their first transmembrane domain, which biochemically resembles a LS except that it is not cleaved. However, proteins without LS can also be secreted by unconventional mechanisms, for example: interleukin, galectin [35]. The process by which such secretory proteins access to the cell exterior is termed unconventional protein secretion (UPS). In plants, even 50% of secreted proteins can be UPS dependent [36]. It has also observed that LS are extremely heterogeneous and that many prokaryotic and eukaryotic LS are functionally interchangeable even between different species [37,38]. Based on these observations, LS from different species can be used to express a gene of interest in defined host cell system. Here, many attempts have been applied to identify potent LS that can be fused to recombinant proteins, in order to improve their secretion efficiency [38]. For such a purpose, it was demonstrated that a native LS is not necessarily the most effective one [39,40]. For targeting to the plasma membrane, proteins have additional sequences (e.g. membrane spanning regions, stop transfer sequences, GPI anchors, etc.) that allow them to attach initially to the ER membrane. As membrane material “flows” from the ER to the Golgi and finally the plasma membrane where it remains attached to a leaflet of the cell membrane. GPI anchor plays an important role in delivering the attached membrane protein to the plasma membrane. GPI is a glycolipid that can be attached to the Cterminus of a protein during post-translational modification. The basic structure of a GPI-anchored protein consists of phosphatidylinositol linked to an unusual non-N-acetyl glucosamine, which, in turn, is linked to three mannose residues followed by an ethanolamine covalently linked to the protein via an amide linkage (EtNP-6Man α1-2Man α1-6Man α1-4GlcN α1-6myoinositol-phospholipid). Depending on the species and functional context, there may exist variations in the side chain associated with the glycan core [10]. Studies on GPI-anchored proteins demonstrated that these proteins could form oligomeric clusters on the cell surface [10]. However, GPI-anchored proteins can exist in different forms depending on the context and the tissue in which they are expressed. Alternate splicing can cause the same protein to exhibit transmembrane, soluble, or GPI-anchored forms; for example, neural cell adhesion molecule (NCAM) can exist in its GPIanchored and soluble form when expressed in muscles; whereas, it take up a transmembrane form instead of the soluble form in brain [10]. GPI-linked proteins are thought to be preferentially located in lipid rafts: areas of the membrane rich in sphingolipids and acylated proteins, as well as cholesterol, suggesting a high level of organization within plasma membrane microdomains, which can serve as a sorting station for a number of cell signaling molecules, thereby functioning as a reaction center. In an early study, fluorescence microscopy of labeled human FOLR1-GPI in the Chinese hamster ovary (CHO) cells showed a diffuse and uniform distribution of the clusters at the outer surface of the intact cell plasma membranes [41]. Once at the cell surface, GPIanchored proteins exhibit a rich diversity of dynamic behaviors in terms of their diffusion, organization, and interactions with other membrane-resident proteins. The chemistry of the fatty acid chain of the GPI anchor is a prerequisite for nanoclustering to occur. Mutation of enzymes affecting lipid remodeling of the GPI anchor, inhibiting the replacement of short and unsaturated acyl chains with long saturated ones, leads to disrupted nanoclustering at the plasma membrane. In addition, the chemistry of the GPI anchor, particularly the nature of the lipid moiety, is extremely important for organizing into cholesterol-sensitive nanoclusters at the plasma membrane [10]. Proteins containing a GPI anchor play key roles in a wide variety of biological processes such as cell signaling and cell adhesion [10]. This has implications in health and disease. For examples, impairment of GPI anchoring is implicated in a large number of diseases, such as the formation of the scrapie form of the prion protein, the causative agent for Creutzfeldt-Jakob disease, and the paroxysmal nocturnal hemoglobinuria is caused by the absence of GPI on the membrane [10]. The role of GPI anchoring is necessary for embryonic development, and its perturbation is the cause of several neurological disorders, abnormal cell growth in yeast, and the survival of many protozoan parasites [10]. Based on these observations, it is therefore crucial to check the localization of the clustering form of GPI-anchored proteins at the cell surface. In the present study, the results obtained from “ghost” of HEK 293 clones confirm the localization of the biosensors as clusters (dots) on the outer surface of the cell plasma membrane of the pmeLUC1 and pmeLUC2 transfected HEK293 cells. These results are in concordance with those obtained for human FOLR1-GPI in CHO cells [41]. Our results showed then the efficiency of the GPI of the human FOLR1 for the expression of the pmeLUC1 and pmeLUC2 biosensors at the outer surface of the cell plasma membrane in HEK293 cells.
In summary, the pmeLUC probe was targeted and expressed on the plasma membrane, with the catalytic site facing the extracellular milieu has some advantages over methods so far available: 1) this topology enables pmeLUC probe to measure ATP increases owing to transient release in the pericellular space of the plasma membrane; 2) the genetic manipulation may allow to measure with this technique ATP levels in vivo. One of the drawbacks of this technique is the need for transfection, which poses a limit to the cell types that may be investigated by this mean. Successful transfection is influenced by many factors: the choice of the transfection method, cell type, health and viability of the cell line, degree of confluency, quality and quantity of the nucleic acid used, etc. HEK293 cells were the ones of choice for transfection because these cells have been widely used in cell biology research for many years due to their reliable growth and propensity for transfection. For our research work, just like Pellegatti [31,32], our choice was with HEK293 cells for transfection. Another stable clones generated by these authors were CT26-pmeLUC cells [31]. As there was no significant difference regarding the structure between our pmeLUC2 probe (without LS of FOLR1 and with GFP instead of myc tag) and that obtained by Pellegatti et al. our pmeLUC2 probe would also work with the CT26 cell lineage for transfection. Also, it would be consequently possible to transform primary cells with our pmeLUC2 probe and engraft them in living mice with successfully across cell passages as observed by Pellegatti et al. [31,32]. In the present work, the use of GFP coupled with Fluc in our pmeLUC2 probe allowed the direct control of transfection process as well as localization of expressed Fluc on the plasma membrane of live cells via the confocal fluorescence microscopy analysis, enabling real-time monitoring of dynamics in situ, and eliminating therefore the potential fixation artifacts, laborious and time-consuming of immunofluorescence analysis needed with myc tag [31]. Indeed, in cell and molecular biology, the GFP gene is frequently used as a reporter of expression [42]. It has been used in modified forms to make biosensors, and many animals have been created that express GFP, which demonstrates a proof of concept that a gene can be expressed throughout a given organism, in selected organs, or in cells of interest. GFP can be introduced into animals or other species through transgenic techniques, and maintained in their genome and that of their offspring. To date, GFP has been expressed in many species, including bacteria, yeasts, fungi, fish and mammals, including in human cells. In the present work, better quality images of the reporter of expression of GFP gene would be obtained by using the 4’, 6-diamidino-2-phenylindole (DAPI) counter staining [43]. In any way, our pmeLUC2 dual-function reporter construct described here takes advantage of the characteristics of both GFP and Fluc for the measurement of pericellular ATP. The combining both reporter genes: GFP and Fluc into a single gene could provide additional tools for the analysis of cancer cells in vivo and ex vivo [44]. Such a dualfunction reporter gene was created and the single encoded protein was shown to be fluorescent and bioluminescent [45]. The GFP portion of the protein allowed for analyses of single living cells expressing the chimeric protein within a population by fluorescence microscopy, and the luciferase activity could be detected from the same living cells. HEK293-pmeLUC cells were among the first stable clones that Pellegatti et al. generated, and in which the pmeLUC probe was extensively validated in vitro as well as in vivo [30,31]. As there was no significant difference regarding the structure between our pmeLUC2 probe (without LS of FOLR1 and with GFP instead of myc tag) and that obtained by Pellegatti et al., it would be consequently possible to inject our stable clones of HEK293- pmeLUC2 in living mice to quantify ATP in vivo and could detect therefore extracellular ATP levels in healthy tissues (<5 μM) as well as high extracellular ATP concentration at tumor sites.
Conclusion
In conclusion, in the present work, the presence of the immobilized firefly luciferase (Fluc) on the cell plasma membrane of the pmeLUC2 transfected HEK293 cells was confirmed by (a) the presence of GFP expression seen only on the surface of the pmeLUC2 transfected HEK293 cells as many dots (clusters) (see in Figure 2D), compared with that seen throughout the positive control of free GFP: vector 9 transfected HEK 293 cells (see in Figure 2C); and (b) the detection and measurement of the luminescence emission from the pmeLUC2 transfected HEK293 “ghost” (dead cell in which the outline remain visible, but without the cytosol as well as other cytoplasmic structures or stainable nucleus obtained after the cell lysis). Our pmeLUC2 dual-function reporter construct appears as suited, sensitive, and reliable biosensors for the local real-time measurement of extracellular ATP concentration in the pericellular space in live mammalian cells. The method used for the construction of our pmeLUC2 probe may pave the way for new strategies applicable to rational pmeLUC design. Its use in live cells and organisms, especially for identifying a new pathway for ATP secretion as a signaling molecule, promise to further expand its utility.
References
- North RA, Verkhratsky A. Purinergic transmission in the central nervous system. Pflugers Arch. 2006;452(5):479-85.
- Pankratov Y, Lalo U, Verkhratsky A, et al. Vesicular release of ATP at central synapses. Pflugers Arch. 2006;452:589-97.
- Burnstock G. Physiology and pathology of purinergic neurotransmission. Physiol Rev. 2007; 87(2):659-797.
- Abbracchis MP, Burnstock G, Verkhratsky A, et al. Purinergic signaling in the nervous system: an overview. Trends Neurosci. 2009;32(1):19-29.
- Burnstock G. Purinergic signalling and disorders of the central nervous system. Nat Rev Drug Discov. 2008;7(7):575-90.
- Burnstock G, Verkhratsky A. Evolution origins of the purinergic signalling system. Acta Acta Physiol. 2009;195(4):415-47.
- Picher M, Burch LH, Boucher RC. Metabolism of P2 receptor agonists in human airways: implications for mucociliary clearance and cystic fibrosis. J Biol Chem. 2004;279(19):20234-41.
- Zimmerman H. Ectonucleotidases in the nervous system. Novartis Found. Symp. 2006; 113-28.
- Fitz JG. Regulation of cellular ATP release. Trans Am Clin Climatol Ass. 2007;118:199-208.
- Saha S, Anilkumar AA, Mayor S. GPI-anchored protein organization and dynamics at the cell surface. J Lipid Res. 2016;57(2):159-75.
- Serna L. A simple method for discriminating between cell membrane and cytosolic proteins. New Phytol. 2005;165(3):947-52.
- Carrea G, Bovara R, Mazzola G. Bioluminescent continuos-flow assay of adenosine 5?-triphosphate using firefly luciferase immobilized on nylon tubes. Anal Chem. 1986;58(2):331-3.
- Carrea G, Bovara R., Girotti S, et al. Continuos-flow bioluminescent determination of ATP in platelets using firefly luciferase immobilized on epoxy methacrylate. J. Biolum. Chemilum. 1989;3(1):7-11.
- Goddard JM, Hotchkiss JH. Polymer surface modification for the attachment of bioactive compounds. Prog Polym Sci. 2007;32(7):698-725.
- Wu H, Liang Y, Shi J, et al. Enhanced stability of catalase covalently immobilized on functionalized titania submicrospheres. Mater Sci Eng C Mater Biol Appl. 2013;33(3):1438-45.
- Naylor LH. Reporter gene technology: the future looks bright. Biochem Pharmacol. 1999;58(5):749-57.
- Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509-44.
- Wilson T, Hastings JW. Annu Rev Cell Dev Biol. 1998;14197-230.
- Niswender KD, Blackman SD, Rohde L, et al. Quantitative imaging of green fluorescent protein in cultured cells: comparison of microscopic techniques, use in fusion proteins and detection limits. J Microsc. 1995;180:109-16.
- Cubitt AB, Heim R, Adams SR, et al. Understanding improving and using green fluorescent proteins. Trends Biochem Sci. 1995;20(11):448-55.
- Li X, Zhao X, Fang Y, et al. Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem.1998;273(52):34970-75.
- DeLuca MA. Ed. Methods in Enzymology. Academic Press. New York. 1978
- Thomson JF, Hayes LS, Lloyd DB. Modulation of firefly luciferase stability and impact on studies of gene regulation. Gene 1991;103(3):171-77.
- TaniyamaY, Tachibana K, Hiraoka K, et al. Development of safe and efficient novel nonviral gene transfer using ultrasound: enhancement of transfection efficiency of naked plasmid DNA in skeletal muscle. Gene Ther. 2002;9(6):372-80.
- Harrison PT, Dalziel RG, Ditchfield NA, et al. Neuronal-specific and nerve growth factor-inducible expression directed by the preprotachykinin-A promoter delivered by an adeno-associated virus vector. Neuroscience 1999;94(3):997-1003.
- Greer LF, Szalay AA. Imaging of light emission from the expression of luciferases in living cells and organisms: a review. Luminescence 2002;17(1):43-74.
- Ignowski JM, Schaffer DV. Kinetic analysis and modeling of firefly luciferase as a quantitative reporter gene in live mammalian cells. Biotechnol Bioeng. 2004;86(7):827-34.
- Liu H, Jiang Y, Luo Y, et al. A simple and rapid determination of ATP, ADP and AMP concentrations in pericarp tissue of litchi fruit by high performance liquid chromatography. Food Technol Biotechnol. 2006;44(4):531-34.
- von Papen M, Gambaryan S, Schutz C, et al. Determination of ATP and ADP secretion from human and mouse platelets by an HPLC assay. Transfus Med Hemother. 2013;40(2):109-16.
- Bhatt DP, Chen X, Geiger JD, et al. A sensitive HPLC-based method to quantify adenine nucleotides in primary astrocyte cell cultures. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;889-890:110-15.
- Pellegatti P, Falzoni S, Pinton P, et al. A novel recombinant plasma membrane-targeted luciferase reveals a new pathway for ATP secretion. Mol Bio Cell. 2005;16(8):3659-65.
- Pellegatti P, Raffaghello L, Bianchi G, et al. Increased level of extracellular ATP at tumor sites: in vivo imaging with plasma membrane luciferase. PLos ONE 2008;3(7):e2599.
- Kapp K, Schrempf S, Lemberg MK, et al. Post-targeting functions of signal peptides. Austin (TX): Landes Bioscience. 2009.
- Blobel G, Dbberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975;67(3):835-51.
- Nickel W, Seedorf M. Unconventional mechanisms of protein transport to the cell surface of eukaryotic cells. Annu Rev Cell Dev Biol. 2008;24:287-308.
- Agrawal GK, Jwa NS, Lebrun MH, et al. Plant secretome: unlocking secrets of the secreted proteins. Proteomics.2010;10(4):799-827.
- von Heijne, G. Signal sequences: the limits of variation. J Mol Biol. 1985;184(1):99-105.
- Kober L, Zehe C, Bode j. Optimized signal peptides for the development of high expressing CHO cell lines. Biotechnol Bioeng. 2013;110(4):1164-73.
- Hesketh J, Ravneberg H, Gjerdrum C, et al. Protein expression system. 2005.
- Young R, Rance J. Mammalian expression vector with a highly efficient secretory signal sequence. 2008.
- Mayor S, Maxfield FR. Insolubility and redistribution of GPI-anchored proteins at the cell surface after detergent treatment. Mol Biol Cell 1995;6(7):929-44.
- Phillips GJ. Green fluorescent protein: a bright idea for study of bacterial protein localization. FEMS Microbiol Lett. 2001;204(1):9-18.
- Kapuscinski J. DAPI: a DNA-specific fluorescent probe. Biotech Histochem. 1995;70(5):220-33.
- Contag, CH, Jenkins D, Contag PR, et al. Use of reporter genes for optical measurements of neoplastic disease in vivo. Neoplasia. 2000;2(1-2):41-52.
- Day RN, Kawecki M, BerryD. Dual-function reporter protein for analysis of gene expression in living cells. Biotechniques 1998;25(5):848-56.