Research Article - Biomedical Research (2017) Volume 28, Issue 4
How many mcr-1-harbouring bacteria were spreading geographically?
Kwang Seung Park1#, Jung Hun Lee1#, Moonhee Park1,2, Kwan Soo Ko3 and Sang Hee Lee1*1Department of Biological Sciences, National Leading Research Laboratory of Drug Resistance Proteomics, Myongji University, Myongjiro, Yongin, Gyeonggido, Republic of Korea
2Division of DNA Analysis, Seoul institute, National Forensic Service, Jiyangro, Yangcheongu, Seoul, Republic of Korea
3Department of Molecular Cell Biology, Sungkyunkwan University School of Medicine, Suwon, Republic of Korea
#These authors contributed equally to this work
- *Corresponding Author:
- Sang Hee Lee
Department of Biological Sciences
Myongji University, Republic of Korea
Accepted date: September 14, 2016
Abstract
Colistin is widely used as an antibiotic of last resort for treating infections caused by multidrug-resistant gram-negative bacteria such as carbapenemase-producing Enterobacteriaceae. Recently, the emergence of plasmid-mediated (horizontally-transferable) colistin resistance (mcr-1) has become a great challenge to global public health. The mcr-1 gene was detected in ESBL (Extended Spectrum β-Lactamase)- producing and/or carbapenemase-producing Enterobacteriaceae. Therefore, there is a huge risk of the emergence of pan-drug-resistant gram-negative bacteria. In this paper we discuss the epidemiological analyses of mcr-1 positive Enterobacteriaceae and structural analyses of PmrC that was recently identified as a protein associated with colistin resistance.
Keywords
Plasmid-mediated colistin resistance, Mcr-1, Epidemiology, Dissemination, PmrC
Introduction
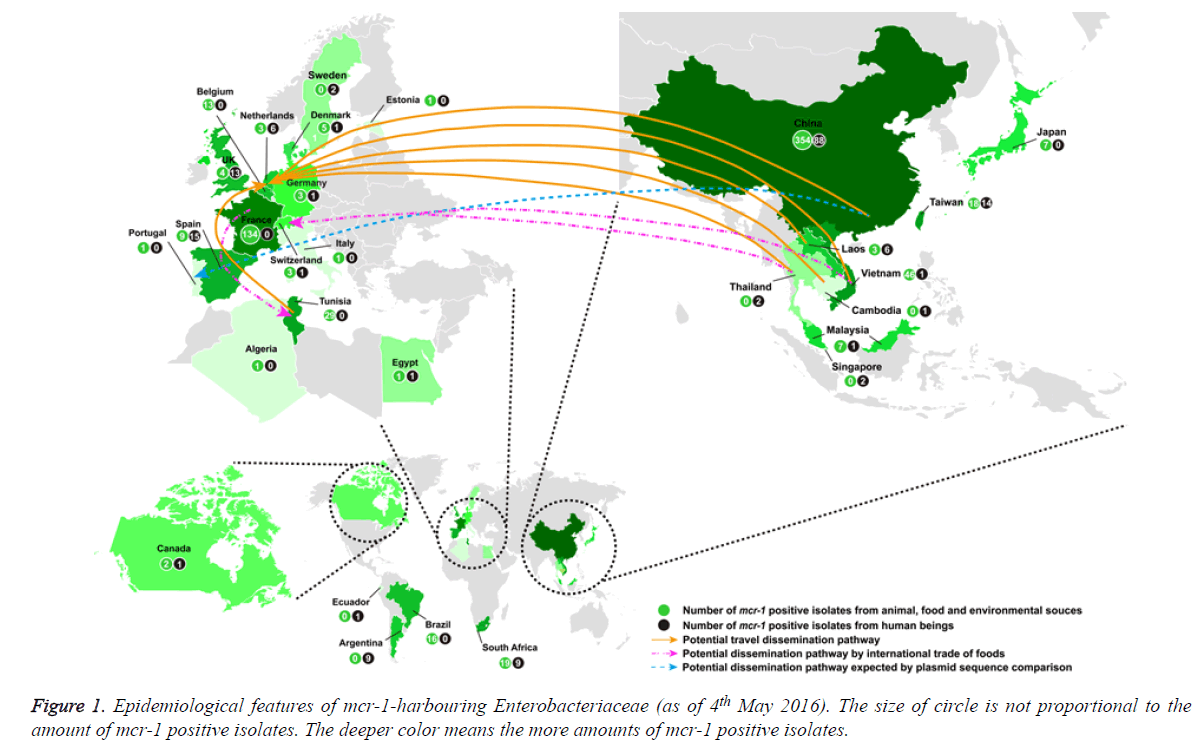
If antibiotic resistant pathogens remain unchecked, it is estimated that by 2050 the global mortality attributed to antibiotic-resistant bacterial infections will soar to 10 million, at a cost of over $100 trillion (http://amr-review.org/). The spread of carbapenemase-producing Enterobacteriaceae is a significant threat to public health. For serious infections caused by carbapenemase-producing Enterobacteriaceae, the treatment options are restricted and invariably rely on tigecycline and colistin [1]. Therefore, the global increase in carbapenemase-producing Enterobacteriaceae has resulted in increased use of colistin with the inevitable risk of emerging resistance. Colistin resistance has involved chromosomal mutations but has never been reported via horizontal gene transfer. However, a plasmid-mediated (horizontally-transferable) colistin resistance (mcr-1) gene was recently reported in China [1] and subsequently detected in Asia (Vietnam, Laos, Thailand, Cambodia, Malaysia, Singapore, Taiwan and Japan), Europe (The Netherlands, Germany, Belgium, Switzerland, France, Denmark, United Kingdom, Spain, Italy, Sweden and Portugal), Africa (Algeria, Egypt, South Africa and Tunisia), and America (Canada, Argentina and Brazil) (Figure 1) [2-47]. We also found two Escherichia coli isolates harbouring mcr-1 gene (GenBank accession no. KU886144 from a human being in Ecuador and GenBank accession no. KU743383 from a pig slurry in Estonia) in the public NCBI database. To investigate how many mcr-1 positive Enterobacteriaceae have been spreading globally, we analysed all these findings that were searched from the following databases PubMed, Medline, Embase, NCBI, and Google Scholar as of 4th May 2016. The analysis results showed the following important aspects: (i) the mcr-1-harboring bacteria had spread to most continents; (ii) four further studies are needed to fight against plasmid-mediated (horizontally-transferable) colistin resistance, particularly in pan-drug-resistant gram-negative bacteria.
Recent global dissemination of mcr-1-harbouring Enterobacteriaceae
As of 4th May 2016, 863 mcr-1 positive Enterobacteriaceae (Escherichia coli, Klebsiella pneumoniae, Salmonella enterica, Enterobacter aerogenes, Enterobacter cloacae, human gut microorganisms and so on) were detected globally and mcr-1- harbouring bacteria have mainly spread in Asia (n=550) and Europe (n=224) (Figure 1) [1-42]. Because in most studies mcr-1 carriers have been identified from random sample collections, the reliable prevalence of mcr-1 positive isolates is not known. In America, only 29 isolates (or 60 in Africa) were detected in animal and/or human being [11,19,21,43-47].
550 (63.7%) of 863 mcr-1 positive Enterobacteriaceae were detected in Asia [1,9,13,15-17,20,21,27-32,35-41] [26.0% in Europe [2-8,10,12,14,17,22-26,33,34,42], 7.0% in Africa [11,21,43,44,46] and 3.3% in America [19,45,47]. mcr-1 positive isolates were distributed to adjacent countries in Asia (or Europe), suggesting the easy spread by a potential travel dissemination pathway as well as a possible dissemination pathway through international trade of foods.
Of note, the spread between Asia and Europe might be allowed by a potential travel dissemination pathway (from China, Vietnam, Laos, Thailand and Cambodia to Netherlands), a potential dissemination pathway by international trade of food (fresh vegetables; from Thailand and Vietnam to Switzerland) or a potential dissemination pathway speculated by the sequence comparison of the isolated plasmids (from China to Portugal) (Figure 1) [4,33,42]. The mcr-1 dissemination between Europe and North Africa might be allowed by a potential travel dissemination pathway (from Tunisia to Netherlands) or a potential dissemination pathway through international trade of foods (chickens, from France to Tunisia) (Figure 1) [4,11].
Scope for further studies
First, mcr-1 positive isolates from human beings were detected after the dissemination of New Delhi Metallo-β-lactamase-1 (NDM-1) positive Enterobacteriaceae that were susceptible to tigecycline and colistin. The limitation of colistin use in treatment of infection via ESBL-producing/mcr-1-harbouring gram-negative bacteria [4,7,10-12,14,19,29,30,35,38] or carbapenemase-producing/mcr-1-harbouring Gram-negative bacteria [9,24,35] may increase tigecycline use and then the possibility of emergence of tigecycline resistance mechanisms other than an efflux pump. To prevent the emergence of a pandrug resistance in gram-negative bacteria, the continued monitoring of colistin and/or tigecycline resistance and their underlining mechanisms in human, animal, food and environmental sources have to be required.
Second, 162 (18.8%) of 863 mcr-1 positive Enterobacteriaceae were investigated about plasmids associated with mcr-1 gene and harboured a low variety of plasmids (IncI2, IncHI1, IncHI2, IncP, IncFI and IncX4) [4,9-13,18,19,23,24,30,32,33,35,38,41]. To clarify the diversity of the plasmid backbones spreading mcr-1 gene within the remaining 701 isolates or isolates detected in the future, additional studies about the plasmids (or the mobile elements) are needed. Like chromosomal localization of the commonly plasmid-borne qnrB (plasmid-mediated quinolone resistance) genes, the chromosomal location of mcr-1 gene associated with a mobile element such as ISApl1 may be observed in the near future. In addition to the plasmid itself, the mobile element alone can represent the transfer mechanism of mcr-1 into Enterobacteriaceae.
Finally, mcr-1 gene was not detected in 803 (48.2%) of 1,666 colistin resistant Enterobacteriaceae [1-44,46,47], suggesting that they might potentially harbour other colistin-resistant mechanism (probably, new groups of MCR) or even novel mcr-1 alleles. The pmrC gene was recently identified as a gene associated with colistin resistance by our group [48]. Homology modelling of MCR-1 and PmrC were performed by an automated homology modelling approach using LptA (PDB ID 4KAY) and EptC (PDB ID 4TN0) as templates in Swiss- Model (http://swissmodel.expasy.org/) program. Although PmrC showed 29% amino acid sequence identity to the MCR-1 sequence, the Root Mean Square Deviation (RMSD) of the Cα trace between PmrC and MCR-1 models was 1.0 Å, supporting that PmrC structure was quite similar to MCR-1 structure as described in the supplementary appendix (Figure S1, available as supplementary material). Structure-based alignment of PmrC, MCR-1, LptA and EptC revealed that key residues identified as to be important to the catalytic activity of LptA and EptC are conserved in PmrC and MCR-1 (Figure S2, available as Supplementary Material). Taken together, the PmrC predicted structure is consistent with a lipid A phosphoethanolamine transferase, which was functionally confirmed by lipid A analysis as previously described [48]. Therefore, the PmrC may probably be another group of MCR. However, further studies are needed to confirm the hypothesis that PmrC may be a new group of MCR in addition to MCR-1 group and continuously monitor another group of MCR.
Supplementary Data
Figures S1 and S2 are available as Supplementary Materials.
Conflict of Interests
The authors declare that they have no competing interests.
Acknowledgements
This work was supported by research grants from the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (No. 2011-0027928 and No. 2016R1C1B2010308) and Marine Biotechnology Program (20150581, Development of Technology for Biohydrogen Production using Hyperthermophilic Archaea) Funded by Ministry of Oceans and Fisheries in Republic of Korea.
References
- Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect Dis 2016; 16: 161-168.
- Hasman H, Hammerum AM, Hansen F, Hendriksen RS, Olesen B, Agersø Y, Zankari E, Leekitcharoenphon P, Stegger M, Kaas RS, Cavaco LM, Hansen DS, Aarestrup FM, Skov RL. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat. Euro Surveill 2015; 20: 30085.
- Anjum MF, Duggett NA, AbuOun M, Randall L, Nunez-Garcia J, Ellis RJ, Rogers J, Horton R, Brena C, Williamson S, Martelli F, Davies R, Teale C. Colistin resistance in Salmonella and Escherichia coli isolates from a pig farm in Great Britain. J Antimicrob Chemother 2016; 71: 2306-2313.
- Arcilla MS, van Hattem JM, Matamoros S, Melles DC, Penders J. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 2016; 16: 147-149.
- Bakterie resistent mot sista behandlingsalternativet funnen. Bacteria resistant to the last treatment option found. Press Stockholm Folkhalsomyndigheten 2016; 10.
- Battisti A. Antibiotic resistance-Italy: colistin, MCR-1, E. coli. Pro Med 2016.
- Cannatelli A, Giani T, Antonelli A, Principe L, Luzzaro F. First detection of the mcr-1 colistin resistance gene in Escherichia coli in Italy. Antimicrob Agents Chemother 2016; 60: 3257-3258.
- Doumith M, Godbole G, Ashton P, Larkin L, Dallman T, Day M, Day M, Muller-Pebody B, Ellington MJ, de Pinna E, Johnson AP, Hopkins KL, Woodford N. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother 2016; 71: 2300-2305.
- Du H, Chen L, Tang YW, Kreiswirth BN. Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect Dis 2016; 16: 287-288.
- Falgenhauer L, Waezsada SE, Yao Y, Imirzalioglu C, Käsbohrer A, Roesler U, Michael GB, Schwarz S, Werner G, Kreienbrock L, Chakraborty T. Colistin resistance gene mcr-1 in extended-spectrum ß-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect Dis 2016; 16: 282-283.
- Grami R, Mansour W, Mehri W, Bouallegue O, Boujaâfar N. Impact of food animal trade on the spread of mcr-1-mediated colistin resistance, Tunisia. Euro Surveill 2016; 21: 30144.
- Haenni M, Poirel L, Kieffer N, Chatre P, Saras E. Co-occurrence of extended spectrum β lactamase and MCR-1 encoding genes on plasmids. Lancet Infect Dis 2016; 16: 281-282.
- Hu Y, Liu F, Lin IY, Gao GF, Zhu B. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 2016; 16: 146-147.
- Kluytmans-van den Bergh MF, Huizinga P, Bonten MJ, Bos M, De Bruyne K. Presence of mcr-1-positive Enterobacteriaceae in retail chicken meat but not in humans in the Netherlands since 2009. Euro Surveill 2016; 21: 30149.
- Kuo SC, Huang WC, Shiau YR, Cheng MF, Lauderdale TL. Colistin resistance gene mcr-1 in Escherichia coli isolates from humans and retail meats, Taiwan. J Antimicrob Chemother 2016; 71: 2327-2329.
- Li A, Yang Y, Miao M, Chavda KD, Mediavilla JR, Xie X, Feng P, Tang YW, Kreiswirth BN, Chen L, Du H. Complete sequences of mcr-1-harboring plasmids from extended- spectrum-β-lactamase-and carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 2016; 60: 4351-4354.
- Malhotra-Kumar S, Xavier BB, Das AJ, Lammens C, Butaye P. Colistin resistance gene mcr-1 harboured on a multidrug resistant plasmid. Lancet Infect Dis 2016; 16: 283-284.
- Malhotra-Kumar S, Xavier BB, Das AJ, Lammens C, Hoang HT. Colistin-resistant Escherichia coli harbouring mcr-1 isolated from food animals in Hanoi, Vietnam. Lancet Infect Dis 2016; 16: 286-287.
- Mulvey MR, Mataseje LF, Robertson J, Nash JH, Boerlin P. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 2016; 16: 289-290.
- Nguyen NT, Nguyen HM, Nguyen CV, Nguyen TV, Nguyen MT, Thai HQ, Ho MH, Thwaites G, Ngo HT, Baker S, Carrique-Mas J. Use of colistin and other critical antimicrobials on pig and chicken farms in southern Vietnam and their association with resistance in commensal Escherichia coli bacteria. Appl Environ Microbiol 2016; 82: 3727-3735.
- Olaitan AO, Chabou S, Okdah L, Morand S, Rolain JM. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 2016; 16: 147.
- Perrin-Guyomard A, Bruneau M, Houee P, Deleurme K, Legrandois P. Prevalence of mcr-1 in commensal Escherichia coli from French livestock, 2007 to 2014. Euro Surveill 2016; 21: 30135.
- Petrillo M, Angers-Loustau A, Kreysa J. Possible genetic events producing colistin resistance gene mcr-1. Lancet Infect Dis 2016; 16: 280.
- Poirel L, Kieffer N, Liassine N, Thanh D, Nordmann P. Plasmid-mediated carbapenem and colistin resistance in a clinical isolate of Escherichia coli. Lancet Infect Dis 2016; 16: 281.
- Prim N, Rivera A, Rodriguez-Navarro J, Espanol M, Turbau M. Detection of mcr-1 colistin resistance gene in polyclonal Escherichia coli isolates in Barcelona, Spain, 2012 to 2015. Euro Surveill 2016; 21: 30183.
- Quesada A, Ugarte-Ruiz M, Rocio Iglesias M, Concepcion Porrero M, Martinez R, Florez-Cuadrado D, Campos MJ, Garcia M, Piriz S, Saez JL, Dominguez L. Detection of plasmid mediated colistin resistance (MCR-1) in Escherichia coli and Salmonella enterica isolated from poultry and swine in Spain. Res Vet Sci 2016; 105: 134-135.
- Ruppe E, Le Chatelier E, Pons N, Andremont A, Ehrlich SD. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 2016; 16: 290-291.
- Shen Z, Wang Y, Shen Y, Shen J, Wu C. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect Dis 2016; 16: 293.
- Stoesser N, Mathers AJ, Moore CE, Day NP, Crook DW. Colistin resistance gene mcr-1 and pHNSHP45 plasmid in human isolates of Escherichia coli and Klebsiella pneumoniae. Lancet Infect Dis 2016; 16: 285-286.
- Suzuki S, Ohnishi M, Kawanishi M, Akiba M, Kuroda M. Investigation of a plasmid genome database for colistin-resistance gene mcr-1. Lancet Infect Dis 2016; 16: 284-285.
- Teo JQM, Cai Y, Lim TP, Tan TT, Kwa ALH. Carbapenem resistance in Gram-negative bacteria: the not-so-little problem in the little red dot. Microorganisms 2016; 4: 13.
- Pham TD, Thanh TH, Nguyen TNT, Chung TH, Wick RR, Thwaites GE, Baker S, Holt KE. Inducible colistin resistance via a disrupted plasmid-borne mcr-1 gene in a 2008 Vietnamese Shigella sonnei isolate. J Antimicrob Chemother 2016; 71: 2314-2317.
- Tse H, Yuen KY. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 2016; 16: 145-146.
- Webb HE, Granier SA, Marault M, Millemann Y, den Bakker HC. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 2016; 16: 144-145.
- Yao X, Doi Y, Zeng L, Lv L, Liu JH. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis 2016; 16: 288-289.
- Ye H, Li Y, Li Z, Gao R, Zhang H. Diversified mcr-1-harbouring plasmid reservoirs confer resistance to colistin in human gut microbiota. MBio 2016; 7: e00177.
- Yu CY, Ang GY, Chin PS, Ngeow YF, Yin WF. Emergence of mcr-1-mediated colistin resistance in Escherichia coli in Malaysia. Int J Antimicrob Agents 2016; 47: 504-505.
- Zeng KJ, Doi Y, Patil S, Huang X, Tian GB. Emergence of the plasmid-mediated mcr-1 gene in colistin-resistant Enterobacter aerogenes and Enterobacter cloacae. Antimicrob Agents Chemother 2016; 60: 3862-3863.
- Zhang R, Huang Y, Chan EW, Zhou H, Chen S. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 2016; 16: 291-292.
- Zhang XF, Doi Y, Huang X, Li HY, Zhong LL, Zeng KJ, Zhang YF, Patil S, Tian GB. Possible transmission of mcr-1–harboring Escherichia coli between companion animals and human. Emerg Infect Dis 2016; 22: 1679-1681.
- Zhi C, Lv L, Yu LF, Doi Y, Liu JH. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 2016; 16: 292-293.
- Zurfuh K, Poirel L, Nordmann P, Nuesch-Inderbinen M, Hachler H, Stephan R. Occurrence of the plasmid-borne mcr-1 colistin resistance gene in ESBL-producing Enterobacteriaceae in river water and imported vegetable samples in Switzerland. Antimicrob Agents Chemother 2016; 60: 2594-2595.
- Coetzee J, Corcoran C, Prentice E, Moodley M, Mendelson M, Poirel L, Nordmann P, Brink AJ. Emergence of plasmid-mediated colistin resistance (MCR-1) among Escherichia coli isolated from South African patients. S Afr Med J 2016; 106: 449-450.
- Elnahriry SS, Khalifa HO, Soliman AM, Ahmed AM, Moustafa AH, Shimamoto T, Shimamoto T. Emergence of plasmid-mediated colistin resistance gene, mcr-1, in a clinical Escherichia coli isolate from Egypt. Antimicrob Agents Chemother 2016; 60: 32449-3250.
- Fernandes MR, Moura Q, Sartori L, Silva KC, Cunha MP, Esposito F, Lopes R, Otutumi LK, Gonçalves DD, Dropa M, Matte MH, Monte DF, Landgraf M, Francisco GR, Bueno MF, de Oliveira Garcia D, Knobl T, Moreno AM, Lincopan N. Silent dissemination of colistin-resistant Escherichia coli in South America could contribute to the global spread of the mcr-1 gene. Euro Surveill 2016; 21: 30214.
- Khalifa HO, Ahmed AM, Oreiby AF, Eid AM, Shimamoto T. Characterisation of the plasmid-mediated colistin resistance gene mcr-1 in Escherichia coli isolated from animals in Egypt. Int J Antimicrob Agents 2016; 47: 413-414.
- Rapoport M, Faccone D, Pasteran F, Ceriana P, Albornoz E, Petroni A, Corso A. Mcr-1-mediated colistin resistance in human infections caused by Escherichia coli: First description in Latin America. Antimicrob Agents Chemother 2016; 60: 4412-4413.
- Park YK, Lee JY, Ko KS. Transcriptomic analysis of colistin susceptible and colistin resistant isolates identifies genes associated with colistin resistance in Acinetobacter baumannii. Clin Microbiol Infect 2015; 21: e1-7.