- Biomedical Research (2012) Volume 23, Issue 1
Host-microbial interaction in the mammalian intestine and their metabolic role inside
Dipendra Raj Pandeya1,2, Roshan D’Souza1, Md. Mashiar Rahman1, Shahina Akhter1,Hyeon-Jin Kim3, Seong-Tshool Hong1*
1Laboratory of Genetics, Department of Microbiology and Immunology, Institute of Medical Science, Chonbuk National University Medical School, Chonju, Chonbuk 561-712, South Korea
2Department of Biochemistry, Nepal Army Institute of Health Sciences, Sanobharayng, Kathmandu, Nepal
3JINIS BDRD Institute, JINIS Biopharmaceuticals Co, 3700 Wilshire Blvd, Los Angeles, California, USA
- Corresponding Author:
- Seong-Tshool Hong
Department of Microbiology, Medical school
Chonbuk national university
Chonju, Chonbuk 561-712
South Korea.
Accepted date: November 16 2011
Abstract
The mammalian gut has coevolved over millions of years with a vast consortium of microbes, which were physically, intimately and densely associated with our body. From birth, this population is in continuous and intimate contact with intestinal tissues. Recent results indicate that indigenous bacteria play a crucial inductive role in gut development during early postnatal life. These findings have revealed that the mammalian intestine is poised for interaction with its prokaryotic partners, which are essential for its normal development. During their coevolution, the bacterial microbiota has established multiple mechanisms to influence the eukaryotic host, generally in a beneficial fashion, and maintain their stable niche. The prokaryotic genomes of the human microbiota encode a spectrum of metabolic capabilities beyond that of the host genome, making the microbiota an integral component of human physiology. Gaining a fuller understanding of both partners in the normal gutmicrobiota interaction may shed light on how the relationship can go awry and contribute to a spectrum of immune, inflammatory, and metabolic disorders and may reveal mechanisms by which this relationship could be manipulated toward therapeutic ends. This review provides a brief overview of this exciting, emerging field.
Keywords
Microbiota, postnatal, coevolution, eukaryotic, prokaryotic, metabolic
Introduction
Prokaryotic organisms can exist in intimate and continuous contact with members of the eukaryotic kingdom. The implications of this statement reflect an emerging theme in the life sciences that has recently come to the forefront of our general view of multicellular plants and animals – that microbes may affect our biology in profound and perhaps previously unsuspected ways. It may be surprising to learn that the human gastrointestinal tract is home to 1014 bacterial organisms [1]. In fact, there are more bacteria in the gut than there are somatic cells in the body. These resident bacteria are referred to as commensal microbiota and their arrival during the first few postnatal days’ sets up a symbiotic association that is necessary and crucial to normal physiology. The complex and dynamic ecosystem of indigenous microbes residing in the intestine may collectively be referred to as the intestinal microbiota. This lifelong association is essential to host pathogen defence and plays an important role in nutrient uptake and metabolism [2]. All mammals are born sterile and immediately after birth, they are initiated into a lifelong process of colonization by foreign microorganisms that inhabit most environmentally exposed surfaces (such as the skin, mouth, gut and vagina) [3]. From that moment on, humans become and remain colonised by microbes. Nearly every surface of mammals that is exposed to the environment is inhabited by commensal bacteria. There is no better example of such a surface than the colon, which contains an astounding number of bacteria, whereas less has been reported from upper gastro-intestinal tract habitats [4]. The thousands of bacteria, fungi and other microbes that live in our gut are essential contributors to our good health. Colonization and the presence of microbiota is important to the development, function and maintenance of a healthy gastrointestinal tract. [5-7].
Over the past five years, studies have highlighted some key aspects of the mammalian host-gut microbial relationship. Gut microbiota could now be considered as a “microbial organ” placed within a host organism. In addition to the obvious role of the intestine in the digestion and absorption of nutrients, the human gastrointestinal tract contains a diverse collection of microorganisms, residing mostly in the colon. As a whole, the microorganisms that live inside humans are estimated to outnumber human cells by a factor of ten. The microbiome represents overall more than 100 times the human genome [8]. Therefore, the gut microbiota and its microbiome provide us with genetic and metabolic attributes, sparing us from the need to evolve solely by our own. Accumulating evidence indicates that the gut microbiota is instrumental in the control of host energy metabolism. These findings open the way to better understand how the gut microbiota and the factors that influence its distribution and constituent microorganisms are controlled and how they interact with the host organism. The present review is intended as an overview of the recent findings in relevant research fields.
Acquisition and establishment of the microbiota
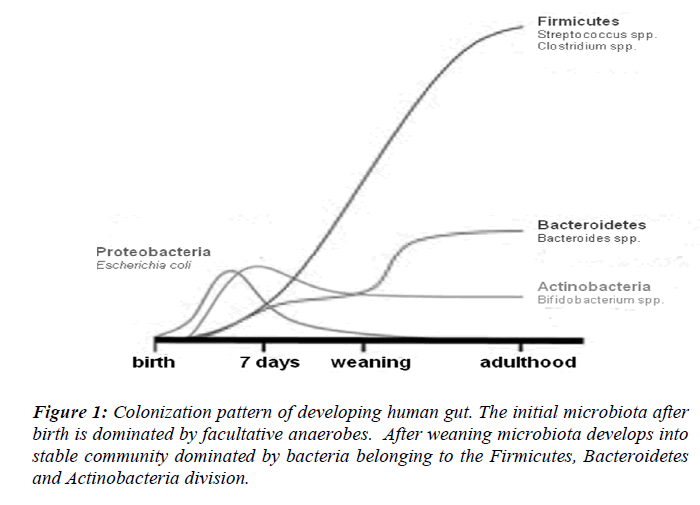
Normal colonization of the sterile newborn intestine is a complex process. Bacteria start colonizing the sterile infant gut within hours after birth followed by a bacterial succession until an adult microbiota has been established post weaning. Upon passage through the birth canal, infats are exposed to a complex microbial population [9]. Evidence that the immediate contact with microbes during birth can affect the development of the intestinal microbiota comes from the fact that the intestinal microbiota of infants and the vaginal microbiota of their mothers show similarities [10]. The gastrointestinal tract is therefore first colonized by facultative anaerobes that lower the redox potential and thus permit growth of strict anaerobes, which normally appear in large numbers during the first week of life [11]. Neonates are quickly colonized by facultative anaerobes (Escherichia coli and Streptococcus), reaching concentrations of 108 to 1010/g of feces within 1–2 days; anaerobic microorganisms do not become established until the second month of life [12] (Fig. 1). In two large studies the dominant bacterial groups in the infant GI-tract were found to be Proteobacteria, Bacteroidetes, Firmicutes, Actinobacteria and Verrucomicrobia [13]. Seemingly, the gut microbiota develops in a chaotic progression during the first months of life depending on bacterial exposure from the surrounding environment. At 6 months of age the human faecal microbiota is dominated by Bacteroidetes and Firmicutes, common occurrence of Verrucomicrobia, and very low abundance of Proteobacteria and aerobic Gram-negative bacteria in general [14]. Predominance of Firmicutes and Bacteroidetes in mammals has been found in several large-scale 16S rRNA sequence-based studies. More than 80% of the identified phylotypes belong to these two phyla in human gut biopsies and faecal samples from a wide range of mammalian species [15,16]. Four hundred to 1000 phylotypes, roughly corresponding to bacterial species, have been estimated to inhabit a healthy human intestine by 16S rRNA cloning and sequencing [17].
It is presumed that this initial colonization is involved in shaping the composition of the gut microbiota through adulthood. For instance, a few studies have shown that kinship seems to be involved in determining the composition of the gut microbiota. Ley et al. [18] have shown that, in mice, the microbiota of offspring is closely related to that of their mothers. The faecal microbiota of adult human individuals is unique and highly stable through time [19,20], and the composition is at least to some extent determined by host genetics [19,21]. Numerous factors govern microbiota stability and shifts (succession changes) in populations. These include intestinal pH, microbial interactions, environmental temperature, physiologic factors, peristalsis, bile acids, host secretions, immune responses, drug therapy, and bacterial mucosal receptors. Both external and host factors control which ingested bacteria will be established in the intestine, and the order of succession of the colonizing strains is of major importance.
Functions of the gut microbiota
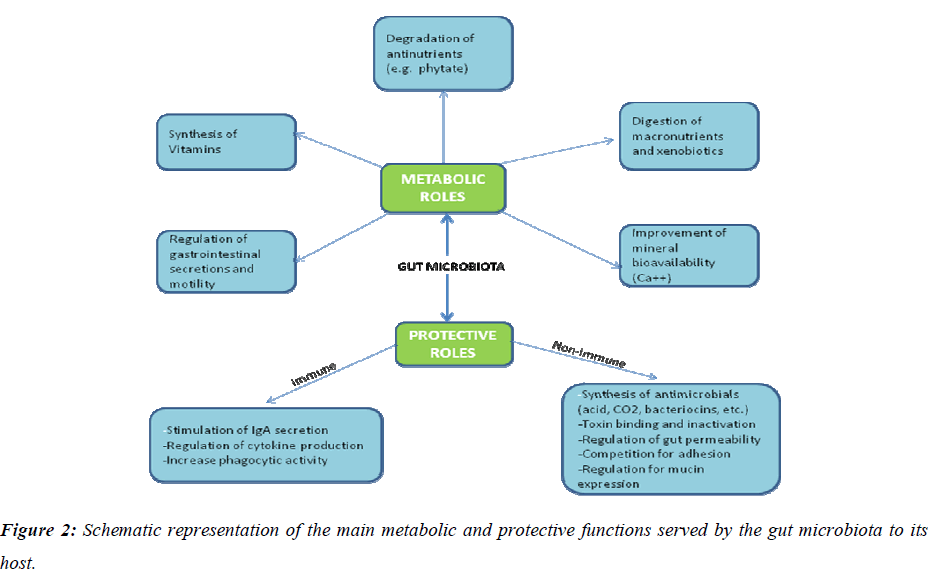
Deciphering biological features of a taxonomically complex and ecologically dynamic microbial community is a challenging issue in gut microbiome research. Recent studies have shown that the complement of gut bacteria varies among individuals, but specific data linking the bacteria present to their functions in human physiology have been lacking. In a recent report, a multidisciplinary approach to link the functions of the trillions of microbial gut bacteria has been described. Germ-free and gnotobiotic mice, [22], pig [23] and zebrafish [24] provide simplified model ecosystems that allow detailed evaluation of functions of colonized microbiota or microbes and the corresponding host responses in vivo, [25] as well as their impact on various host physiologies [26] and more broadly on health. All these data showed that intestinal microbes play a pivotal role in maintaining human health and wellbeing (Fig.2).
Molecular analysis of intestinal microbiota
The extent of diversity of the microbiota is a fundamental question in intestinal microbial ecology. The intestinal microbiota remains incompletely characterized and its diversity poorly defined [15]. For years, the 10-metrelong human intestinal tract was like a dark tunnel. Some light had been shed on it by culturing bacteria from the faeces, but the darkness was overwhelming, because about 70 to 90 percent of the bacteria cannot be cultivated in laboratory dishes. These uncultured bacteria remained completely unknown. Microbiologists knew that trillions of microbes live in the gut, but they had no idea which ones.
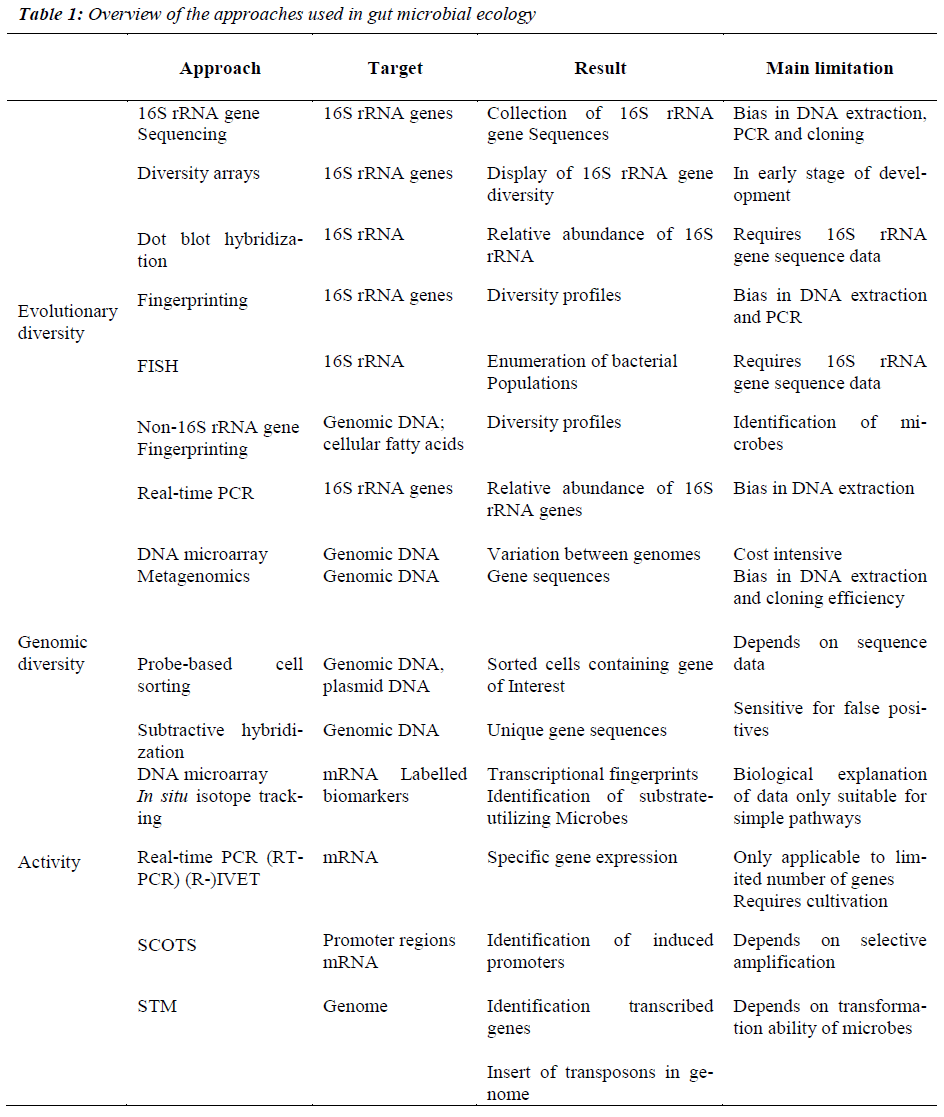
Cultivation-based techniques traditionally were used to study GI bacteria. However, cultivation-based approaches are limited by 3 major factors. First, one can culture only those organisms for which nutritional and growth requirements are known. Second, phenotypic criteria do not reliably enable phylogenetic identification. Third, cultivation techniques are, by design, tedious and impractical for studying ecosystems characterized by extensive microbial diversity. A pivotal technological advance has been the circumvention of a major impediment to microbial research inability to culture the majority of microbes in the gut. The progress made in molecular microbial ecology over the last 10 years has allowed the exploration of the colonic microbiota in humans. This has been achieved by new molecular techniques, notably metagenomics and compositional sequencing [7], which have enabled the study of mixed communities of microbes in the gut, and revealed greater diversity than previously imagined. This situation is changing rapidly, however, thanks to new high throughput technologies and matching software, explained Willem de Vos from Wageningen University and Helsinki University. For the last two or three years, it has become possible to find out which ‘species’or ‘phylotypes’[ 15] of bacteria live in our body, by analyzing the genes for 16S rRNA’s with a phylogenetic microarray, facilitate for the first time our ability to analyze microbiota in depth and in an efficient manner (Table 1). However, this method provides little information about microbial functions, making it difficult to understand microbiota- derived contributions to disease pathologies. The complexity of gut microbial ecology and its impact on health can be better understood by deciphering the genetic information contained in the complete microbial population. Approaches based on genome sequences offer powerful insights into the physiological potential of microbes. Genomics and metagenomics will provide a wealth of information and this requires continuous developments and improvements in high-throughput sequencing technologies and bioinformatics. These approaches have been widely applied to study bacterial communities in the GI tract [27]. The established metabolic profiling approach has a powerful capacity for detecting various metabolites originating from microorganisms that are commonly found in mammals. Microbial community structure can also be analyzed via fingerprinting techniques, whereas dot-blot hybridization, fluorescent in situ hybridization (FISH), or quantitative PCR that target known taxa can measure the abundance of particular microbes. Emerging approaches, such as those based on functional genes and their expression and the combined use of stable isotopes and biomarkers are also being developed and optimized to study metabolic activities of groups or individual organisms in situ.
Several analytical techniques, including high resolution nuclear magnetic resonance (NMR) spectroscopy45 and various gas liquid chromatography–mass spectrometry (GC–Ms, LC–Ms) techniques [28] are currently used to generate spectral profiles from which information pertaining to pathophysiology can be extracted. Studies of gut microbiota interactions with metabolic phenotypes (socalled functional metagenomics) are now possible through the use of proton nuclear magnetic resonance (1H NMR) based profiling of fecal, urine or other extracts. Early results in this area that tried to correlated microbiota and probiotic supplementation-induced changes in its composition are promising [29].
Diversity of GI -tract microflora in different sites of the digestive tract
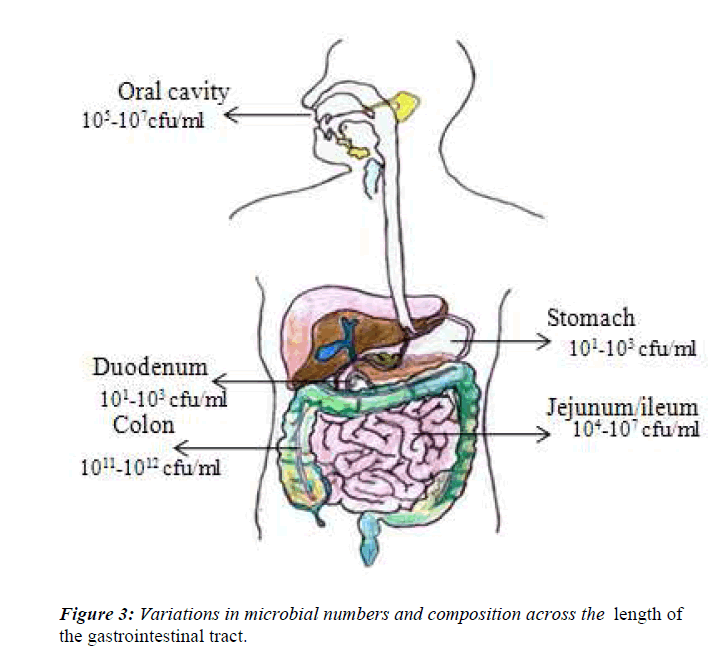
It is now recognized that all external body surfaces have a normal resident bacterial flora, and this includes the digestive tract. During the last 25 years there has been a massive increase in our knowledge about the gut flora at different sites in the digestive tract [30]. This has occurred as a result of improved anaerobic culture techniques, the recognition of the limitations on the data and attempts to moderate those limitations. Accumulating data indicate that the indigenous intestinal microbiota are an essential component of human physiology. Bacteria are present throughout the human gastrointestinal tract, but the number and spectrum of microbes vary considerably due to differences in pH, presence of immune factors and digestive enzymes, and transit time in different parts of the intestine (Fig.3). Although the present of microbiota on the skin, in the respiratory tract, in the vagina, but majority are located in the colon of gastrointestinal tract, which has major impact on the health. The adult digestive tract can be viewed as a succession of different ecosystem. Indeed, microbial populations are unevenly divided throughout the digestive tract due to a mosaic of ecological conditions (e.g. Physicochemical, nutritional condition and transit time), resulting in important between- organ differences. The stomach represents a split in the microbial continuum of the digestive tract because of its extremely acidic pH, as most of the microorganisms ingested are destroyed before reaching the duodenum. This makes it difficult to distinguish the exact proportions of resident versus transient stomach bacteria [1]. However, the conditions are less extreme in the small intestine and large intestine, where the pH varies between 6.5-7.5 in the small intestine [31] from the duodenum to the ileum, and between 5.5-7.0 in the large intestine from proximal to its distal part [32].
Relatively few studies have been published on the diversity of the microflora in the human intestine [33,34] or on the flora associated with the mucosa [35]. Problems are associated with sampling at various sites in the GI-tract, because invasive techniques such as intubation or collection of material during operation have to be employed to obtain samples of intestinal contents. Invasive sampling techniques can usually not be used for sampling of healthy subjects. In general, the faecal flora seems to represent the colonic flora. However, there is a great variation in microflora compared with the upper sites of the digestive tract.
Host-microbial symbiosis
Normal intestinal microbiota are characterized as a complex collection and balance of microorganisms that normally inhabit the healthy GI tract. The indigenous bacteria sometimes have been classified as potentially harmful or health-promoting. Most of them, however, are part of the normal commensal flora. This term indicates a relationship between organisms of 2 or more different species in which 1species derives benefits from the association while the other(s) remain(s) unharmed or unaffected. Currently, the relationship between intestinal bacteria and the host is referred to as host–microbe cross-talk, implying peaceful coexistence and mutual benefit.
Cross talk between the host and commensal microbes has been an area of intense investigation during the past decade, but the mechanisms by which bacteria in the gut influence host physiology and by which host physiology influences bacteria in the gut remain largely unknown.
Cooperative interactions between eukaryotes and prokaryotes are well known. In these symbiotic relationships, the microbe profits by acquisition of a stable temperature, oxygen and nutrient supply. Eukaryotic hosts may gain extended metabolic/digestive ability and benefit from competitive exclusion of harmful microbes. The present body of knowledge regarding host-commensal cross talk has been derived mainly from studies conducted using germ-free or gnotobiotic animals. In addition, the promising potential of using selected strains of intestinal microbes as probiotics has spawned research elucidating the effects of specific probiotic bacteria in vitro and in vivo [36]. However, data from these studies should be interpreted cautiously for a number of reasons. Commensal microbes constitute a dynamic ecosystem characterized by interaction between members of the microbiota and the host. Thus, the effects of selected commensal microbes to host physiology may in part depend on microbe-microbe interactions, which are difficult to recapitulate and investigate in an experimental setting. Initial microbial colonization in the neonatal period coincides with structural and functional maturation of the intestinal immune system, and the expression of many of the immune molecules involved in recognition of microbial structures is developmentally regulated. In addition to their homeostatic function in later life, indigenous microbes appear to play an important developmental role in early infancy. After the intestinal microbiota has been established, its composition remains relatively stable. The host is tolerant toward its indigenous microbiota, that is, immune responses toward commensal bacteria are local and noninflammatory in nature. It is likely that host responses to resident commensal bacteria differ from those elicited toward initial colonizers in early life or toward nonpathogenic microbes that do not belong to the indigenous microbiota, such as probiotics.
Metabolic potential of the intestinal microbiota
The intestinal microbiota may also play an important role in human health by means of its metabolic potential. The human gut microbiota is a metabolic organ having a coding capacity that exceeds that of the liver by a factor 100 [37] and whose composition is determined by a dynamic process of selection and competition. The gut microbiome enlarges the genome of the host and enhances the host’s metabolic potential [37]. Indeed, it is estimated that the collection of all microbial genomes in the gut comprises between 2 million and 4 million genes, which is 70–140 times more than that of their host [38]. This ‘microbiome’ encompasses all genes that are responsible for numerous processes such as substrate breakdown, protein synthesis, biomass production, production of signaling molecules, anti- microbial compounds and it encodes biochemical pathways that humans have not evolved [39]. The intestinal microbiota can therefore be regarded as a separate organ within the human host that is capable of even more conversions than the human liver, and we can view ourselves as a composite of human cells and bacteria and our genetic landscape as a ‘metagenome,’ an amalgam of genes embedded in our genome and in the genomes of all our microbial partners [7]. Therefore, from the metabolic point of view it would be more correct to describe human as ‘superorganisms’ that is a human/ microbes hybrid. Through its immense metabolic capabilities, the gut microbiota contributes to human physiology by transforming complex nutrients, such as dietary fiber or intestinal mucins that otherwise would be lost to the human host, into simple sugars, short-chain fatty acids and other nutrients that can be absorbed [40]. Furthermore, the microbiota produces some essential vitamins including vitamin K, vitamin B12 and folic acid, contributes to intestinal bile acid metabolism and recirculation.
In contrast to the oxidative and conjugative nature of liver metabolism, which generates hydrophilic high molecular weight biotransformation products, the metabolic nature of the gut microbial community in an anaerobic environment is mainly reductive and hydrolytic, generating nonpolar low molecular weight byproducts [41]. Additionally, the intestinal microbiota also interferes with the human biotransformation process through the enterohepatic circulation of xenobiotic compounds. Compounds that have been absorbed in the intestine and subsequently detoxified are usually conjugated with polar groups (glucuronic acid, glycine, sulfate, glutathion and taurine) in the epithelium or liver. Such metabolites may enter the blood stream prior to excretion in the urine, but depending on the compound a considerable fraction may also enter again into the intestine via secretion with the bile [42]. Once released in the intestinal lumen, these conjugates may be hydrolyzed again by bacterial enzymes such as β-glucuronidases, sulfatases and glucosidases.
Mechanisms for microbial regulation of host metabolism
To understand how the gut microbiota affects host physiology, recently it has been investigated that how the gut microbiota regulates the metabolome and transcriptome in germ-free and conventionally raised mice. Metabolomic analysis revealed that the gut microbiota affects several important metabolic processes including energy metabolism, amino acid, and lipid metabolism. The serum metabolome was associated with increased hepatic transcription of genes involved in proteolysis, energy, and xenometabolism. Surprisingly, we detected increased levels of neurotransmitters in serum of conventionally raised animals, which suggests that the gut microbiota may affect animal behavior. Taken together, these results suggest that variations in an individual’s gut microbiota may have profound effects on host metabolism and physiology and will be an important factor when considering personalized medicine.
Metabolite production by gut bacteria
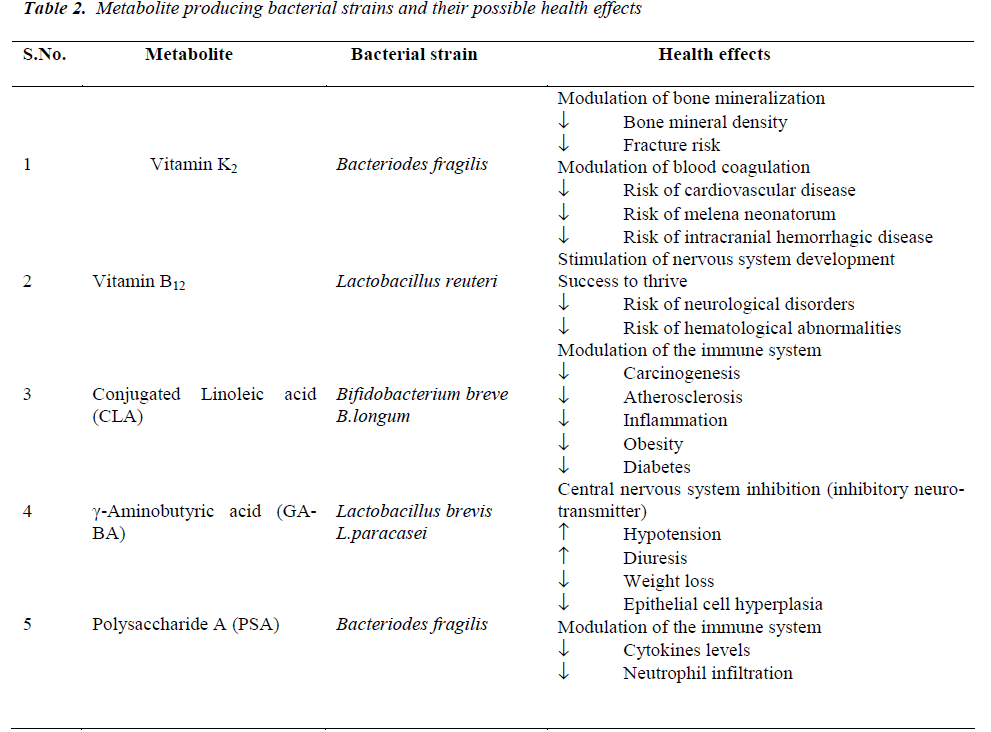
The human enteric microbiota can exert beneficial health effects through the production of bacterial metabolites or ‘pharmabiotics’, most often small molecules which interact with ‘intelligent communication’ systems in the body including those which are immune, endocrine and neuronal- based. Commensal bacteria have been shown to synthesise vitamins that are essential for human survival such as vitamins K and B [43], polyunsaturated fatty acids (PUFA) such as conjugated a-linolenic acid (CALA) and CLA, SCFA, neuroactive compounds such as GABA and histamine [44], PSA and a variety of other proteins, peptides and nucleotides with immunomodulatory and anti-inflammatory properties [43]. The effects of some of these compounds on human health are briefly reviewed below and summarised in (Table 2).
Nutritional benefits of the gut flora
Nutrients from digested bacterial cells may be of greatest benefit to a mammalian host when they are made available in the stomach or intestine. In addition to products of hydrolysis of the macromolecular constituents of their cells, lysed microbial cells are also sources of vitamins and other cofactors [45]. Germfree rats are known to require vitamin K in their diets, while conventional rats do not [45]. Likewise, germfree rats and animals of certain other species require in their diets certain B vitamins (e.g. B12, biotin, folic acid, and pantothenate) in concentrations higher than those required by their conventional counterparts [45]. In these cases, the substances may derive predominantly from organisms residing in the cecum and colon, but may come as well from microbial cells growing on epithelial surfaces in the fore- and midguts.
In 1983, Wostmann and colleagues observed that germfree rodents require 30% more calories to maintain their body mass than conventional rodents (possessing their ‘normal’ gut flora) [46]. The potential mechanisms accounting for this observation remained obtuse until recently when seminal studies by Drs. Lora Hooper, Jeffrey Gordon and others using germ-free mice colonized with conventional gut flora or B.thetaiotaomicron suggested that the gut flora contribute to carbohydrate and lipid absorption [47-51]. Sequencing of the B. thetaiotaomicron genome revealed, remarkably, that a majority of this genome is devoted to polysaccharide utilization and, importantly, contains enzymatic capacities lacking in the human genome permitting, for example, the digestion of nutrients otherwise inaccessible to the host [48]. The genome of these bacterial glycophiles, termed a ‘glycobiome’, predicts that they display receptors for complex polysaccharides as well as secrete a vast array of carbohydrate- degrading enzymes into the bacterial periplasm or extracellular fluid [47]. Consistent with the hypothesis that the metabolic capabilities of B. thetaiotaomicron are critical to host nutrition, these organisms are observed to associate with food particles and mucus and to modify their glycan foraging behavior (via differential gene expression) depending on the available nutrient sources [51]. The gut flora also likely regulates fat storage [49].
Finally, microorganisms influence the nutrition of the animal tissues of their host. For example, they may provide certain vitamins, carbon energy, and nitrogen in the form of metabolic end-products and macromolecular precursors. The microbial cells may also compete for available, dietary nutrients with the host's animal cells. Therefore, for these microbial communities, the contributive and competitive nutritional activities are significant to the nutrition of the animal tissues.
Role of the microbiota in host energy metabolism
Microbial communities are characterized by unparalleled complexity. Our increasing technological ability to characterize this complexity will contribute to understanding the ecological processes that drive microbe-eukaryotic interactions. One of the most striking findings that helped to define this mutualistic relationship was the role of microbiota in energy harvest. The role of the gut microbiota in host energy and metabolism is becoming more and more clear and is considered a critical factor, together with lifestyle, involved in energy metabolism and obesity. The gut microbiota increase energy absorption from the gut by direct mechanisms. To examine the relationship between the composition of the gut microbiota and the efficiency of energy harvest, the levels of SCFA, the major fermentation end-products and source of energy for the host, influence the host gene expression in systemic tissue such as the liver (lipogenesis, gluconeogenesis) and adipose tissue (lipogenesis and inflammation) and the energy content of the faeces were used as markers of energy harvesting.
Whether changes in the microbiota are a cause or consequence of obesity [52] remains to be established, but it is clear that the microbiota aid in energy harvesting from our foods. The hypothesis that changes in our microbiota through lifestyle changes and modern medical practice, which might lead to disappearance of certain species within the microbiota [53], species which are intimately involved in human physiology and the disappearance of which leads to modern-day disorders, requires further investigation. But, until the role of the microbiota in obesity is clear, we will have to carefully watch the balance between energy intake and expenditure.
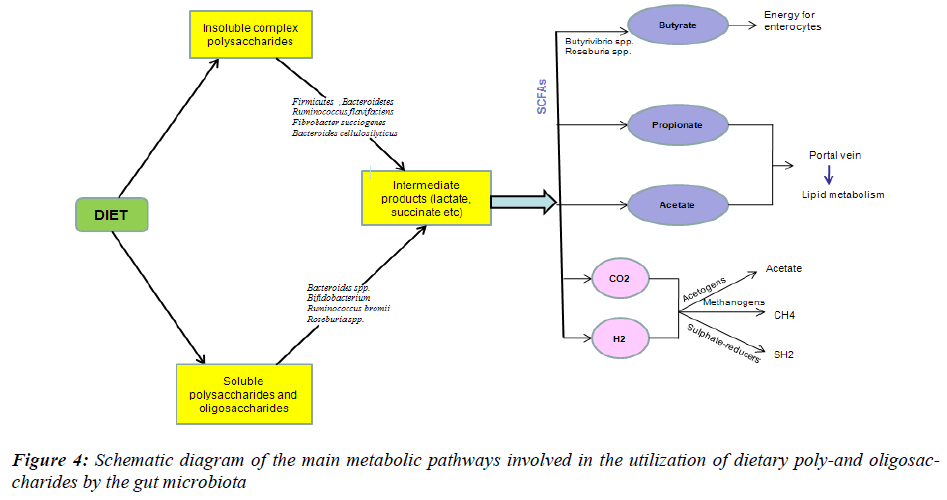
The amount of energy that is stored in the body depends on the balance between energy intake and expenditure: when energy intake exceeds expenditure, excess energy is stored as fat, which leads to weight gain and eventually obesity. To examine the relationship between the composition of the gut microbiota and the efficiency of energy harvest, the levels of SCFA, the major fermentation endproducts and source of energy for the host, and the energy content of the faeces were used as markers of energy harvesting (Fig. 4). Faecal SCFA and energy content were then correlated with the levels of Firmicutes, Bacteroidetes and Actinobacteria in lean, ob/ob and HF-fed mice. Evidence on the role of the gut microbiota on energy harvesting from the diet came from studies performed in germ-free mice [54]. Ba¨ckhed et al. [49] found that conventionally raised mice contained 40% more total body fat and 47% higher gonadal fat content than germ-free mice. Several pathways are proposed to explain that the presence of the gut microbiota drives the increase in fat mass – shown both in animals fed a standard carbohydrate- rich or a fat-rich diet [55,56].
The gut microbiota may also influence energy balance by modifying gene expression of host-related metabolic functions. Angiogenesis, which is primarily involved in distributing nutrients to peripheral tissues, was shown to depend on the gut microbial colonization process. Although capillary network formation was arrested in adult germfree mice, this developmental process restarted and was completed within 10 days after colonization with a complete microbiota harvested from conventionally raised mice, or with Bacteroides thetaiotaomicron. Commensal bacteria, such as B. thetaiotaomicron, have also been shown to induce expression of host monosaccharide transporters in monocolonized mice. This would lead to increasing the absorption of monosaccharides and SCFA and, thereby, promote the novo synthesis of lipids in the liver.
In different studies, when the distal gut microbiota from the normal mice was transplanted into the gnotobiotic mice, there was a 60% increase in body fat within 2 weeks. To clarify possible mechanisms of this effect, the authors showed that the microbiota promoted absorption of monosaccharides from the gut and induced hepatic lipogenesis in the host, responses mediated by 2 proteins: carbohydrate response element-binding protein (ChREBP) and liver sterol response element-binding protein type-1 (SREBP-1) [49], energy extraction from nondigestible food components (via short chain fatty acids (SCFA) production through the fermentation). In an interesting experiment, using genetically modified (fastinginduced adipocyte factor [Fiaf]–knockout) mice, the same authors showed that gut microbes suppress intestinal Fiaf, also known as angiopoietin-like protein 4. Fastinginduced adipocyte factor inhibits lipoprotein lipase activity, thereby catalyzing the release of fatty acids from lipoprotein associated triacylglycerols, which are then taken up by muscle and adipose tissue. In the study, Fiaf suppression resulted in increased lipoprotein lipase activity in adipocytes and promoted storage of calories as fat, leading the authors to postulate that energy regulation by the gut microbiota occurs through at least three interrelated microbial mechanisms: a) fermentation of indigestible dietary polysaccharides to absorbable forms; b) intestinal absorption of monosaccharides and short chain fatty acids with their subsequent conversion to fat within the liver, and c) regulation of host genes that promote deposition of fat in lipocytes [49]. In conclusion, changes in the proportions of the major phyla of the gut microbiota were unrelated to markers of energy harvest which changed over time. These findings suggest that microbial adaptation to diet over time, and perhaps with age, is an important variable in the complex relationship between the composition of the microbiota, energy harvesting capacity and obesity and should be considered in future studies.
Toxic Consequences of Gut Bacterial Metabolism
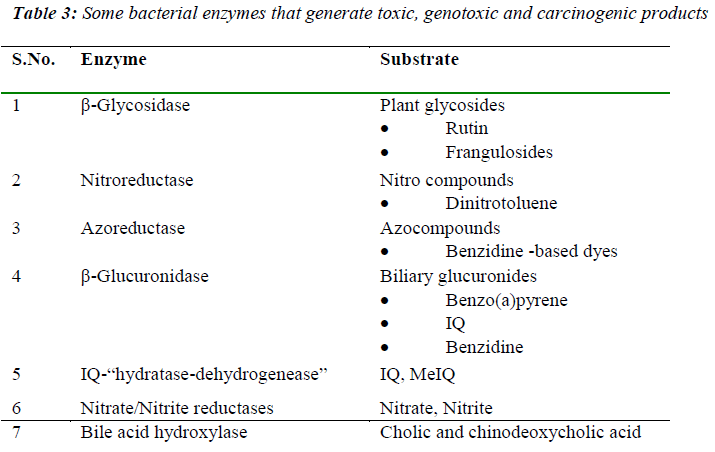
The metabolic activities of the gut microform have also been associated with carcinogenic processes such as tumor promotion (ammonia, secondary bile acids), mutagenesis (fecapenaenes), and carcinogenesis (N-nitroso compounds). The enzymic activities of the gut microflora, towards ingested foreign compounds such as nitroaromatics, azo compounds, and nitrate can have wideranging implications for health, since bacterial metabolism of such compounds can lead to the generation of genotoxic and carcinogenic products. Bacterial enzymes commonly assayed include β-glucuronidase, β-glycosidase, azoreductase, nitroreductase, nitrate reductase, the conversion of pre-carcinogen 2 amino-3-methyl-7Himidazo [4,5-f] quinoline (IQ) to 7-hydroxy-2-amino-3,6- dihydro-3-methyl-7Himidazo[4,5-f]quinoline-7-one (7OHIQ). A list of the major bacterial enzymatic reactions leading to alterations in the toxicity of substrates is given in Table 3.
Future Prospects
The importance of understanding the relationships between humans and their resident microbial populations has been emphasized for over 100 years. The study of symbiosis in the mammalian gastrointestinal tract has been hampered because of the inherent difficulties in analyzing such a complex ecosystem. A basic mechanistic understanding of host–microbial interactions in the mammalian gut will ultimately yield new strategies for the prevention and treatment of some infectious diseases in humans. Analysis of interactions between commensals and their hosts will require the active participation of scientists from many disciplines: cellular microbiologists, immunologists, experts in functional genomics and bioinformatics, applied mathematicians, and individuals from the field of materials science. Obtaining a picture of the molecular underpinnings of symbiotic or commensal relationships should lead to new approaches for the prevention and treatment of infectious diseases. The biochemical signals used by the microbes themselves to effect stable host–microbe and/or microbe–microbe interactions may represent new classes of drugs useful for maintaining our indigenous microbial barriers. Finally, by identifying these microbial signals, we may develop important new insights about the strategies that pathogens employ to gain entrance into, and eventual control of an ecosystem.
Conclusion
The human microflora has a significant impact on health and human disease, far more than ever realized. Different mechanisms have been proposed to explain the link between gut flora and obesity. The future will almost certainly hold a dramatic shift in our thinking about our interactions with the prokaryotic world. Instead of something to be avoided at all costs, perhaps these relationships will eventually be viewed as an integral part of our biology. These developments might ultimately help us to understand the microbial ecology of the GI tract and provide us with insight into the mechanisms underlying GI tract health and disease. Finally, the capacity to enumerate the microbiota represents a first step in understanding molecular contributions of this microbial society to human physiology. To fully define our own metabolic potential, it will be necessary to define the metabolic potential of our microbiota. This effort should include initiation of a systematic effort to sequence the microbiome [25] and to develop methods for monitoring microbial gene expression in vivo (first in gnotobiotic mouse models and later in humans). Over our evolutionary history, components of the intestine’s microbiota have endured a stringent selection to become “master physiologic chemists”: i.e., they have had to develop chemical strategies for regulating nutrient processing in ways that benefit themselves and us. By identifying these host genes and the microbial effectors of their expression/function, we should be able to identify new molecular targets and new chemical strategies for manipulating nutrient processing, uptake, and utilization.
Acknowledgement
This paper was supported by research funds of Chonbuk National University in 2009
References
- Savage DC. Microbial ecology of the gastrointestinaltract. Annu Rev Microbio 1977; 31: 107-133.
- Nicholson JK, Holmes E, Wilson ID. Gut microorganisms,mammalian metabolism and personalizedhealthcare. Nat Rev Microbiol 2005; 3: 431-438.
- Round JL, Mazmanian SK. The gut microbiota shapesintestinal immune responses during health and disease.Nat Rev Immunol 2009; 9: 313-323.
- Andersson AF, Lindberg M, Jakobsson H, BäckhedF, Nyrén P, Engstrand L. Comparative analysis ofhuman gut microbiota by barcoded pyrosequencing.PLoS One 2008; 30: e2836.
- Chu FF, Esworthy RS, Chu PG, et al. Bacteriainducedintestinal cancer in mice with disrupted Gpx1and Gpx2 genes. Cancer Res 2004; 64: 962-928.
- Pull SL, Doherty JM, Mills JC, et al. Activated macrophagesare an adaptive element of the colonic epithelialprogenitor niche necessary for regenerative responsesto injury. Proc Natl Acad Sci U S A 2005;102: 99-104.
- Turnbaugh PJ, Ley RE, Hamady M, et al. The humanmicrobiome project. Nature 2007; 449: 804-810.
- Walker A, Cerdeño-Tárraga A, Bentley S. Faecal matters.Nat Rev Microbiol 2006; 4: 572-573.
- Redondo-Lopez V, Cook RL, Sobel JD. Emergingrole of lactobacilli in the control and maintenance ofthe vaginal bacterial microflora. Rev Infect Dis 1990;12: 856-872.
- Mandar R, Mikelsaar M. Transmission of mother’smicroflora to the newborn at birth. Biol Neonate1996; 69: 30-35.
- Fanaro S, Chierici R, Guerrini P, Vigi V. Intestinalmicroflora in early infancy: composition and development.Acta Paediatr Suppl 2003; 91: 48-55.
- Thompson-Chagoyán OC, Maldonado J, Gil A. Colonizationand impact of disease and other factors on intestinalmicrobiota. Dig Dis Sci. 2007; 52: 2069-2077
- Favier CF, Vaughan EE, De Vos WM, AkkermansAD. Molecular monitoring of succession of bacterialcommunities in human neonates. Appl Environ Microbiol2002; 68: 219-226.
- Palmer C, Bik EM, DiGiulio DB, Relman DA, BrownPO. Development of the human infant intestinal microbiota.PLoS Biol 2007; 5: e177.
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, DethlefsenL, Sargent M, Gill SR, Nelson KE, RelmanDA. Diversity of the human intestinal microbial flora.Science. 2005; 308:1635-1638.
- Ley RE. Obesity and the human microbiome. CurrOpin Gastroenterol 2010; 26:5-11.
- Rajilic-Stojanovic, M, Smidt, H, de Vos, WM. Diversityof the human gastrointestinal tract microbiota revisited.Environ Microbiol 2007; 9: 2125-2136.
- Ley RE, Backhed F, Turnbaugh P, et al. Obesity altersgut microbial ecology. Proc Natl Acad Sci U S A2005; 102:11070-11075.
- Zoetendal EG, Akkermans AD, Akkermans-van VlietWM, de Visser J, de Vos WM. The host genotype affectsthe bacterial community in the human gastrointestinaltract. Microb. Ecol. Health Dis 2001; 13: 129-134
- Jernberg C, Lo¨ fmark S, Edlund C, Jansson JK.Longterm ecological impacts of antibiotic administrationon the human intestinal microbiota. ISME J 2007;1: 56-66.
- Stewart JA, Chadwick VS, Murray A. Investigationsinto the influence of host genetics on the predominanteubacteria in the faecal microflora of children. J MedMicrobiol 2005; 54: 1239-1242.
- Bry L, Falk PG, Midtvedt T, Gordon JI. A model ofhost-microbial interactions in an open mammalianecosystem, Science 1996; 273: 1380-1383.
- Meurens F, Berri M, Siggers RH, Willing BP, SalmonH, Van Kessel AG, Gerdts V. Commensal bacteriaand expression of two major intestinal chemokines,TECK/CCL25 and MEC/CCL28, and their receptors.PLoS One 2007; 7: e677.
- Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafishreveal evolutionarily conserved responses to thegut microbiota. Proc. Natl. Acad. Sci. USA 2004;101: 4596-4601.
- Hooper LV, Wong MH, Thelin A, Hansson L, FalkPG, Gordon JI. Molecular analysis of commensalhost-microbial relationships in the intestine. Science2001; 291: 881-884.
- Dumas ME, Barton RH, Toye A, Cloarec O, BlancherC, Rothwell A, et al. Metabolic profiling reveals acontribution of gut microbiota to fatty liver phenotypein insulin-resistant mice. Proc. Natl. Acad. Sci. USA2006; 103: 12511-12516.
- Zoetendal EG, Collier CT, Koike S, Mackie RI, GaskinsHR. Molecular ecological analysis of the gastrointestinalmicrobiota: a review. J Nutr 2004; 134: 465-472.
- Qiu Y, Su M, LiuY, Chen M, Gu J, Zhang J, Jia W.Application of ethyl chloroformate derivatization forgas chromatography-mass spectrometry based metabonomicprofiling. Anal. Chim. Acta 2007; 583: 277-283.
- Martin FP, Wang Y, Sprenger N, Yap IK, LundstedtT, Lek P, Rezzi S, Ramadan Z, van Bladeren P, FayLB, Kochhar S, Lindon JC, Holmes E, Nicholson JK.Probiotic modulation of symbiotic gut microbial-hostmetabolic interactions in a humanized microbiomemouse model. Mol Syst Biol 2008; 4: 157.
- Drasar BS, Hill MJ. Human Intestinal Flora. AcademicPress, London, U.K. 1974.
- Evans DF, Pye G, Bramley R, Clark AG, Dynson TJ,Hardcastle JD. Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut1988; 29: 1035-1041.
- Cummings JH, Macfarlane GT. The control and consequencesof bacterial fermentation in the human colon.J appl Bacteriol 1991; 70: 443-459.
- Ventura M, Turroni F, Canchaya C, Vaughan EE,O'Toole PW, van Sinderen D. Microbial diversity inthe human intestine and novel insights from metagenomics.Front Biosci. 2009; 14: 3214-3221.
- Booijink CC, Zoetendal EG, Kleerebezem M, de VosWM. Microbial communities in the human small intestine:coupling diversity to metagenomics. FutureMicrobiol 2007; 2: 285-295.
- Peach SL, Tabaqchali S. Some studies of the bacterialflora associated with the mucosa of the human gastrointestinaltract. Nahrung 1984; 28: 627-634.
- Rautava S, Kalliomaki M, Isolauri E: New therapeuticstrategy for combating the increasing burden of allergicdisease: Probiotics-A Nutrition, Allergy, MucosalImmunology and Intestinal Microbiota (NAMI) ResearchGroup report. J Allergy Clin Immunol 2005;116: 31-37.
- Possemiers S, Grootaert C, Vermeiren J, et al. Theintestinal environment in health and disease: recentinsights on the potential of intestinal bacteria to influencehuman health. Curr Pharm Des 2009; 15: 2051-2065.
- Qin J, Li R, Raes J, Arumugam M, Burgdorf S, ManichanhC, et al. A human gut microbial gene catalogueestablished by metagenomic sequencing. Nature 2010;464: 59-65.
- Egert M, de Graaf AA, Smidt H, de Vos WM, VenemaK. Beyond diversity: functional microbiomicsof the human colon. Trends Microbiol 2006; 14: 86-91.
- Volker M, Maria U, David JB. Understanding theExtent and Sources of Variation in Gut MicrobiotaStudies; a Prerequisite for Establishing Associationswith Disease. Diversity 2010; 2: 1085-1096.
- Sousa T, Paterson R, Moore V, Carlsson A, AbrahamssonB, Basit AW. The gastrointestinal microbiotaas a site for the biotransformation of drugs. Int JPharm 2008; 363: 1-25.
- Ilett KF, Tee LBG, Reeves PT, Minchin RF. Metabolismof drugs and other xenobiotics in the gut lumenand wall. Pharmacol Therapeut 1990; 46: 67-93.
- Collado MC, Isolauri E, Salminen S, Sanz Y. Theimpact of probiotic on gut health. Curr Drug Metab2009; 10: 68-78.
- Ross RP, Mills S, Hill C, Fitzgerald GF, Stanton C.Specific metabolite production by gut microbiota as abasis for probiotic function. International Dairy Journal2010; 20: 269-276.
- Savage DC. Gastrointestinal microflora in mammaliannutrition. Annu Rev Nutr 1986; 6: 155-178.
- Wostmann BS, Larkin C, Moriarty A, Bruckner-Kardoss E. Dietary intake, energy metabolism, andexcretory losses of adult male germfree Wistar rats.Lab Anim Sci 1983; 33: 46-50.
- Backhed F, Ley RE, SonnenburgJL, Peterson DA,Gordon JI. Host-bacterial mutualism in the human intestine.Science 2005; 307: 1915-1920.
- Xu J, Bjursell MK, Himrod J, DengS, Carmichael LK,Chiang HC, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science2003; 299: 2074-2076.
- Backhed F, Ding H, Wang T, Hooper LV, Koh GY,Nagy A, et al. The gut microbiota as an environmentalfactor that regulates fat storage. Proc NatlAcad Sci USA 2004; 101: 15718-15723.
- Hooper LV, Wong MH, Thelin A, Hansson L, FalkPG, Gordon JI. Molecular analysis of commensalhost-microbial relationships in the intestine. Science2001; 291: 881-884.
- Sonnenburg JL, Xu J, Leip DD, Chen CH, WestoverBP, Weatherford J, et al. Glycan foraging in vivo byan intestine-adapted bacterial symbiont. Science 2005;307: 1955-1959.
- Backhed F. Changes in intestinal microflora in obesity:cause or consequence? J Pediatr GastroenterolNutr 2009; 48: S56-S57.
- Blaser MJ, Falkow S. What are the consequences ofthe disappearing human microbiota? Nat Rev Microbiol2009; 7: 887-894.
- Ley RE. Obesity and the human microbiome. CurrOpin Gastroenterol. 2010; 26: 5-11.
- Cani PD, Lecourt E, Dewulf EM, Sohet FM, PachikianBD, Naslain D, et al.Gut microbiota fermentationof prebiotics increases satietogenic and incretingut peptide production with consequences for appetitesensation and glucose response after a meal. AmericanJournal of Clinical Nutrition 2009a; 90: 1236-1243.
- Musso G, Gambino R, Cassader M. Gut microbiota asa regulator of energy homeostasis and ectopic fat deposition:mechanisms and implications for metabolicdisorders. Curr Opin Lipidol 2010; 21: 76-83.