Research Article - Biomedical Research (2017) Volume 28, Issue 21
Highly unique and stable biomarkers for diagnosis of Mycobacterium tuberculosis pathogens
Guangxin Yuan and Guangyu Xu*
College of Pharmacy, Beihua University, Jilin, PR China
Accepted date: September 06, 2017
Abstract
Introduction: Tuberculosis (TB) is one of the most extensively spread diseases in developing countries. A recent survey found that the incidence of tuberculosis showed a clear upward trend. Therefore, in order to better control tuberculosis infection, finding fast and accurate method for the early diagnosis is very important. Polymerase Chain Reaction (PCR) is a simple, rapid, sensitive and unique genetic diagnosis technology, but there are still some major problems in its applification for the early diagnosis of TB. With the development of modern computational biology, it has become easier to screen for new TB diagnostic targets.
Materials and Methods: We first screened the genes which are only present in M. tuberculosis by blast technology. We used the keywords “Rv1512” “Rv1513” “Rv1516” “Rv1973” ”Rv1974” ”Rv2646” “Rv2655” “Rv2659” “Rv3119” “Rv3120” “Rv3617” “Rv3738c”to search the publications from 2000 to 2016 in PubMed. Using Epidata3.1, we deleted the repeated and unrelated studies by parallel entry and logical error test. In the end, the specificity of using the genes for the diagnostic screening of clinical tuberculosis specimens was validated by PCR method.
Results: Using clinical tuberculosis specimens, the specificity of the obtained genes were validated by PCR method, and the genes were compared with those of tuberculosis, asthma, pneumonia, and lung cancer with similar early symptoms to those of tuberculosis. It was finally verified that the 3 genes obtained by screening can be used as the biomarkers for the early diagnosis of tuberculosis.
Conclusion: In conclusion we find that these genes can be used as targets for the early diagnosis of tuberculosis.
Keywords
Mycobacterium tuberculosis, Screen, Diagnostic genes, PCR
Introduction
Tuberculosis (TB) is one of the most widespread respiratory infectious diseases [1]. Early symptoms of TB are similar to those of some other diseases, such as lung cancer, asthma, and pneumonia, leading to the delaying of the diagnosis and to the spreading of TB infection [2]. Therefore, more effective, rapid and accurate early diagnostic methods are urgently needed, in order to control the spreading of TB.
There are a lot of TB diagnostic methods, but the traditional diagnostic methods are time-consuming, and some new diagnostic methods, such as proteomics, transcriptomics and metabolomics, and phage mining methods, require high-level technologies although they are specific in the diagnosis of TB, so that the new methods are not conducive to the clinical application. Polymerase Chain Reaction (PCR) is a simple, rapid, sensitive, and specific gene diagnostic technology, and its technical requirements are not so high, but widely used [3]. However, its use is limited to due to lack of reliable biomarkers, which results in false positive and false negative results [4,5].
In this study, our goal was to identify marker genes unique to M. tuberculosis by comparative genomics, and then we validated the unique of the marker genes on clinical M. tuberculosis samples using PCR [6,7]. The targets obtained in this study showed a strong specificity and stability.
Materials and Methods
Ethical statement
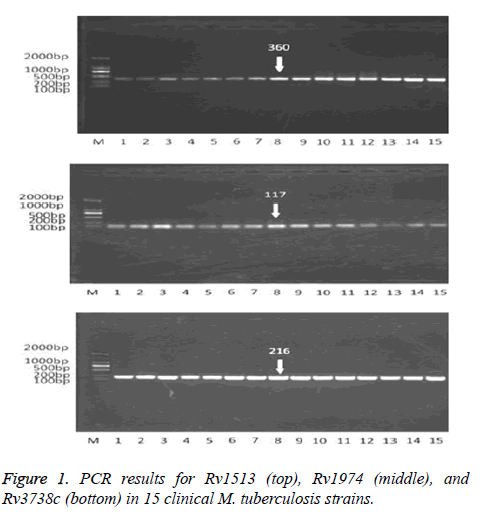
The patients or their close relatives signed on the written informed consents after the study was described and explained to them. The distribution profile of Mycobacterium tuberculosis was evaluated using 15 TB samples (Figure 1) from 105 patients suffering typical measles symptoms, who were admitted to our hospital from 2010 to 2011. The bacterial culture and serological experiments were carried out to verify all of the 15 M. tuberculosis samples with, and then the samples were stored at -20°C for use.
Data collection
Genome sequences of all microorganisms and the standard M. tuberculosis strain H37Rv (Accession numbers: NC_000962) was collected and downloaded from GenBank.
Bibliometric method
We used the keywords “Rv1512” “Rv1513” “Rv1516” “Rv1973” ”Rv1974” ”Rv2646” “Rv2655” “Rv2659” “Rv3119” “Rv3120” “Rv3617” “Rv3738c”to search the publications from 2000 to 2016 in PubMed [8]. Using Epidata3.1, we deleted the repeated and unrelated studies by parallel entry and logical error test.
Screening for genes unique to M. tuberculosis H37Rv with comparative genomics
Search for genes unique for tuberculosis is carried out with BLAST against all microbial genome sequence. Hit sequences were kept at an E-value cut-off of 1E-5 and 50% identity. Candidate sequences were then examined for stability, i.e., whether flanked by transposable elements in each of the M. tuberculosis genomes.
Extraction of genomic DNA from M. tuberculosis H37Rv
DNA was isolated from sputum samples from 15 patients, according to the previously published method [9,10]. Briefly, 5 ml sodium hydroxide (1 mol/L) was added to 500 μL sputum to allow vibrating liquefaction for 30 min.
Samples were pelleted by centrifugation for 5 min at 14000 r/ min, washed with 1.5 mL deionized water. Equal volume of the mixture of chloroform and isopropanol (24:1) was added to the samples, and supernatant was removed by centrifugation.
Ethyl alcohol of two volumes and 1/10 volume of 3 mol/L natrium aceticum were added and mixed well. DNA samples were then pelleted and washed with 70% ethanol.
Finally, DNA is dissolved in 100μL Tris-HCL and 1 × EDTA buffer solution after air drying at room temperature.
Extraction of genomic DNA from asthma, pneumonia, and lung cancer samples
We extracted DNA from the sputum of patients with asthma, pneumonia and lung cancer. We extracted genome DNA of the cell pellet by using QIAamp DNA Mini Kit.
PCR primer design
The primers were designed by Primer Premier 5 software and selective Coding Sequence (CDS) for the conserved sequence design primer was used (Table 1). The pre-amplification fragments covered most of the main amino acids sites of the CDS and were respectively synthesized by Sangon Biotech, LLC.
| Gene | Sequence (5′-3′) | Expected length (bp) |
|---|---|---|
| Rv1513 | TCCGTCTTGCCGATGTT | 360 |
| ATCGGCCTGCAGCATTC | ||
| Rv1974 | GTGGGTGCGCTGCTATGC | 117 |
| GGCGTTGGCGAAGTTGT | ||
| Rv3738c | CCGAGTTGGCCGCTAATCA | 216 |
| CAGCACCGCCGTTGAACA |
Table 1. PCR primer sequences.
PCR conditions
The standard PCR conditions included 30 cycles of 5 min at 94°C for pre-denaturation, 30 s at 94°C for denaturation, 30 s at 55°C for annealing, and 60 s at 72°C for extending. The 30 cycles were followed by a final extension at 72°C for 10 min. The Taq is TaKaRa Taq Kit. The thermal cycler is Thermo Scientific Arktik Thermal Cycler.
Ethical statement
The study was approved by the Ethics Committee of School of Basic Medical Sciences, Jilin University and each patient was consented in a written informed consent form.
Results
Candidate diagnostic markers
All genes of M. tuberculosis H37Rv were compared with all other microbial genes that have been aligned using BLAST (Evalue< e-5, sequence identity>95%). 12 genes unique only to M. tuberculosis, were identified, which are only present in M. tuberculosis and do not have homologous genes in other mycobacterium spp. such as M. avium and M. bovis (Table 2).
| Gene | Gene ID | Function | Reference |
|---|---|---|---|
| Rv1512 | 886461 | Nucleotide-sugar epimerase epiA | -10 |
| Rv1513 | 886464 | Hypothetical protein | |
| Rv1516c | 886455 | Sugar transferase | -11 |
| Rv1973 | 885936 | MCE associated membrane protein | -12 |
| Rv1974 | 885935 | Hypothetical protein | |
| Rv2646 | 887706 | Integrase | -13 |
| Rv2655c | 887388 | phiRv2 prophage protein | |
| Rv2659c | 885098 | phiRv2 prophage integrase | |
| Rv3119 | 888811 | Molybdenum cofactor biosynthesis protein E | |
| Rv3120 | 888828 | Hypothetical protein | |
| Rv3617 | 885769 | Epoxide hydrolase | -14 |
| Rv3738c | 886262 | PPE family protein |
Table 2. Candidate diagnostic target genes of H37Rv.
Screening by bibliometric search
The searches using the bibliometric method were conducted since the candidate diagnostic genes might have been identified previously by other methods. Five genes, Rv1512, Rv1516, Rv1973, Rv2646, and Rv3617, were excluded from the further analysis since they had previously been used as diagnostic targets (Table 2).
Screening for genetic stability
The stability of diagnostic markers is an important factor, and one criterion for the stability is whether the flanking sequences contain transposons, integrons, or other transposable genetic elements [11,12]. Hence, the transposons, integrons, and other movable genetic elements in the flanking sequences within 1500 bp of the candidate diagnostic targets were analyzed in this study. Many transposons were present within 1500 bp of Rv2655 and Rv2659c, so that it might be unstable for these genes to be used as the diagnostic markers. There were anomalous and labile regions within 1500 bp of Rv3119, Rv3120, and Rv3617, so that these genes were not suitable to be used as the biomarkers either. Table 3 shows details about the flanking sequences around the candidate diagnostic genes. Based on this analysis, Rv1513, Rv1974, and Rv3738c had conservative flanking sequences.
| Gene | 1500 bp flanking region | ||
|---|---|---|---|
| tRNA | Transposase | Phage | |
| Rv1513 | |||
| Rv1974 | |||
| Rv3738c | |||
| Rv2655c | + | + | |
| Rv2659c | + | + | |
| Rv3119 | + | ||
| Rv3120 | + | ||
Table 3. Flanking sequences of candidate diagnostic targets in H37Rv.
PCR validation of the biomarkers in clinical strains of M. tuberculosis
The functions of Rv1513 and Rv1973 genes are unclear and Rv3738c is one of the Pro-Pro-Glu (PPE)/Pro-Glu (PE) family members. It was necessary to verify the three candidate diagnostic markers-Rv1513, Rv1973, and Rv3738c-by PCR. DNA from 15 clinical M. tuberculosis strains was extracted and the PCR was performed using the primers shown in Table 1 for the three candidate biomarkers. The PCR results showed that the positive rate of detecting the three genes in clinical isolates was 100%, with a good sensitivity and accuracy (Figure 1).
Validation of TB diagnostic biomarkers with asthma, pneumonia, and lung cancer samples
Early symptoms of TB are similar to those of pneumonia, asthma, and lung cancer. Patients with these diseases suffer coughs and show some shadows in the lung in computed tomography scans, which may make it difficult to differentiate TB from the other diseases. In order to test the specificity of the three diagnostic genes, the diagnostic targets in the samples of asthma, pneumonia, and lung cancer were detected by PCR. All the PCR results from these samples showed negative. And the PCR results of Rv1513, Rv1974, and Rv3738c showed positive in 15 clinical M. tuberculosis strains, as shown in Figure 1. Figure 1 shows the PCR results for Rv1513 (top), Rv1974 (middle), and Rv3738c (bottom) in 15 clinical M. tuberculosis strains.
Discussion
In this study, we searched in the M. tuberculosis H37Rv genome for unique biomarkers that could be used for diagnosis of early TB. Three genes, Rv1513, Rv1976, and Rv3738c, were identified after genome comparison and stability test. The unique of these biomarkers were validated on clinical M. tuberculosis specimens using the PCR [13-15]. Additionally, when tested for samples from asthma, pneumonia, and lung cancer, whose symptoms were considered to be similar to those of TB, the results were negative. All these evidence suggests that the genes are TB-unique diagnostic markers.
Five of the identified 12 genes have already been used in PCR diagnostic methods. These genes account for 42% of the genes identified in our screen, which may suggest that comparative genomics should be a powerful way to identify species-unique markers. These five genes have been used as tuberculosis diagnostic targets only on a small scale, but not as gold standards. The stability of these genes has been not screened, likely leading to a certain amount of false positives in PCR assays for M. tuberculosis detection, although the genes are unique for M. tuberculosis [16].
PCR technology with the right markers can improve the sensitivity and specificity of the detection for infectious diseases, and shorten the time of bacterial detection by traditional culture method, especially with some advantages, including fast, accurate, low in technical requirements and widely used, compared with the new technologies such as proteomics, transcriptome and metabolomics, and phage mining methods [17]. This study is mainly based on the characteristics of rapid and sensitive of PCR technique in vitro amplification, while other molecular biology methods can detect most known gene mutations, gene deletions and chromosome dislocations of M. tuberculosis, so that the detection of unique genes of M. tuberculosis by PCR technique can be one of the most effective and reliable methods for the early diagnosis of tuberculosis. PCR technology is simple and practicable and widely used, but some deficiencies have been also found [18]. For example, the amplification inhibitors, amplification temperature differences as well as the random errors in nucleic acid weight which occurs in the purification process of specimens or nucleic acid may cause false negative results. Furthermore, the cross-contamination between the previous amplification products and extraction products of nucleic acid may cause the false positive results. These are the problems call for a precaution while using this method.
In this study, a 100% positive rate of detecting the three candidate target genes was verified by PCR in a large amount of M. tuberculosis clinical strains. The results showed a good sensitivity and accuracy, suggesting that if they are tested and validated in clinic in large scale samples, the genes may be used as early diagnostic targets of M. tuberculosis in the future. A 100% negative rate of detecting the three candidate target genes was also verified by PCR in asthma, pneumonia, and lung cancer samples because the early symptoms of these diseases are similar to those of TB, demonstrating that the identified genes should be unique to M. tuberculosis and exist only in M. tuberculosis rather than the other pathogens, such as Mycobacterium [19].
Targets used as diagnosis require two characteristics, that is, strong specificity and stability. The genes we identified showed a very strong specificity. In addition to the screening for their specificity, their stability was also examined. To a certain degree, this screening may eliminate false positives while M. tuberculosis is detected. The flanking sequences of the genes for transposons, integrons, and other movable genetic elements were screened to determine the stability of three target genes, Rv1513, Rv1973, and Rv3738c. The three target genes had conservative flanking sequences, so that they can be used as diagnostic targets.
Rv3738c is one of the Pro-Pro-Glu (PPE)/Pro-Glu (PE) family members, and the family is a specific protein family of Mycobacterium involved in the pathogenicity and infection of M. tuberculosis [20]. Genes of the PE/PPE family were once used as diagnostic targets, but were prone to false positive or false negative results because they were screened only for specificity, but not for stability [4]. The Rv3738c gene was screened for both specificity and stability in this study, so that it could be used as a diagnostic target. The functions of Rv1513 and Rv1973 genes are unclear, so it is speculated that they may be unique and necessary genes in the M. tuberculosis family.
Conclusion
Rv1513, Rv1973, and Rv3738c were identified as new and highly unique diagnostic targets for M. tuberculosis following the data mining and screening for stability. Based on the PCR testing, it is believed that Rv1513, Rv1973, and Rv3738c are only present in M. tuberculosis, rather than in other organisms or those to cause non-tuberculosis diseases, so they can be used as diagnostic targets for TB. The diagnostic method is simple and rapid, and can be used for the early diagnosis and largescale infection of TB.
Declaration of Interest
The authors declare that they have no competing interests.
Acknowledgement
This work was supported by the National Natural Science Foundation of China (81401712).
References
- Xu G, Wang J, Guo Y, Bai Y, Yuan G, An L, Han X, Du P, Zhang N, Zheng Y. Analysis of the molecular evolution of the Mycobacterium tuberculosis drug-resistant gene rpoB in Asia using a Bayesian evolutionary method. Clin Lab 2015; 61: 1017-1025.
- Porcel JM, Leung CC, Restrepo MI, Takahashi K, Lee P. Year in review 2012: lung cancer, respiratory infections, tuberculosis, pleural diseases, bronchoscopic intervention and imaging. Respirol 2013; 18: 573-583.
- Pholwat S, Stroup S, Foongladda S, Houpt E. Digital PCR to detect and quantify heteroresistance in drug resistant Mycobacterium tuberculosis. PLoS One 2013, 8: 57238.
- Gomez Pastrana Duran D, Torronteras Santiago R, Caro Mateo P, Lopez Barrio AM, Macias Mardones P, Andres Martin A, Pineda Mantecon M, Navarro Gonzalez J. Effectiveness of smears and cultures in gastric aspirate samples in the diagnosis of tuberculosis. An Esp Pediatr 2000; 53: 405-411.
- Cheng VC, Yew WW, Yuen KY. Molecular diagnostics in tuberculosis. Eur J Clin Microbiol Infect Dis 2005; 24: 711-720.
- Wang TC, Chen FC. The evolutionary landscape of the Mycobacterium tuberculosis genome. Gene 2013; 518: 187-193.
- Bouwman AS, Kennedy SL, Muller R, Stephens RH, Holst M, Caffell AC, Roberts CA, Brown TA. Genotype of a historic strain of Mycobacterium tuberculosis. Proc Natl Acad Sci USA 2012; 109: 18511-18516.
- Ramos JM, Padilla S, Masia M, Gutierrez F. A bibliometric analysis of tuberculosis research indexed in PubMed, 1997-2006. Int J Tuberc Lung Dis 2008; 12: 1461-1468.
- Silva JL, Leite GG, Bastos GM, Lucas BC, Shinohara DK, Takinami JS, Miyata M, Fajardo CM, Luchessi AD, Leite CQ. Plasmid-based controls to detect rpoB mutations in Mycobacterium tuberculosis by quantitative polymerase chain reaction-high-resolution melting. Mem Inst Oswaldo Cruz 2013; 108: 106-109.
- Cao YT, Jia HY DF. Cloning, prokaryotic expression and specificity identification of Mycobacterium tuberculosis Rv1512 gene. J Clin Pulmo Med 2011; 16: 1887-1889.
- Al-Khodari NY, Al-Attiyah R, Mustafa AS. Identification, diagnostic potential and natural expression of immunodominant seroreactive peptides encoded by five Mycobacterium tuberculosis-specific genomic regions. Clin Vaccine Immunol 2010; 18: 477-482.
- Song H, Sandie R, Wang Y, Andrade-Navarro MA, Niederweis M. Identification of outer membrane proteins of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2008; 88: 526-544.
- Ding ZX CN, Zhang XX. Functional study of region of differences in Mycobacterium tuberculosis and its application. J Clin Pulmo Med 2012; 17: 505-507.
- Biswal BK, Garen G, Cherney MM, Garen C, James MN. Cloning, expression, purification, crystallization and preliminary X-ray studies of epoxide hydrolases A and B from Mycobacterium tuberculosis. Acta Crystallogr Sect F Struct Biol Cryst Commun 2006; 62: 136-138.
- Zhuang ZG, Zhang JA, Luo HL, Liu GB, Lu YB, Ge NH, Zheng BY, Li RX, Chen C, Wang X, Liu YQ, Liu FH, Zhou Y, Cai XZ, Chen ZW, Xu JF. The circular RNA of peripheral blood mononuclear cells: Hsa_circ_0005836 as a new diagnostic biomarker and therapeutic target of active pulmonary tuberculosis. Mol Immunol 2017; 90: 264-272.
- Shen L, Wu X, Tan J, Gu M, Teng Y, Wang Z, Yue W. Combined detection of dickkopf-1 subtype classification autoantibodies as biomarkers for the diagnosis and prognosis of non-small cell lung cancer. Onco Targets Ther 2017; 10: 3545-3556.
- Ciric M, Moon CD, Leahy SC, Creevey CJ, Altermann E, Attwood GT, Rakonjac J, Gagic D. Metasecretome-selective phage display approach for mining the functional potential of a rumen microbial community. BMC Genomics 2014; 15: 356.
- Moreira OC, Verly T, Finamore-Araujo P, Gomes SAO, Lopes CM, de Sousa DM, Azevedo LR, da Mota FF, d'Avila-Levy CM, Santos-Mallet JR, Britto C. Development of conventional and real-time multiplex PCR-based assays for estimation of natural infection rates and Trypanosoma cruzi load in triatomine vectors. Parasit Vectors 2017; 10: 404.
- Sharma K, Sharma M, Singh S, Modi M, Sharma A, Ray P, Varma S. Real-time PCR followed by high-resolution melting curve analysis: A rapid and pragmatic approach for screening of multidrug-resistant extrapulmonary tuberculosis. Tuberculosis (Edinb) 2017; 106: 56-61.
- Lew JM, Kapopoulou A, Jones LM, Cole ST. TubercuList-10 years after. Tuberculosis (Edinb) 2011; 91: 1-7.