Research Article - Archives of General Internal Medicine (2017) Volume 1, Issue 2
Global Methicillin-Resistant Staphylococcus Aureus (MRSA) Infections and Current Research Trends
Robert Gallagher1, Noboru Motohashi2, Anuradha Vanam3 and Rao Gollapudi1*
1University of Kansas, Lawrence, KS-66045, USA
2Meiji Pharmaceutical University, Tokyo, Japan
3Sri Venkateswara University, Tirupathi, AP State, India
- *Corresponding Author:
- Rao Gollapudi
University of Kansas
Lawrence, KS-66045, USA
E-mail: gollapudirao@ku.edu
Accepted on April 17, 2017
Citation: Gallagher R, Motohashi N, Vanam A, et al. Global methicillin-resistant Staphylococcus aureus (MRSA) infections and current research trends. Arch Gen Intern Med. 2017;1(2):3-11
Abstract
Methicillin-Resistant Staphylococcus aureus (MRSA), manifested as a primary cause of skin infections, was invasive among healthy adults and children with clinical, epidemiologic, and bacteriologic characteristics distinct from healthcare-associated MRSA (HA-MRSA) resulting in increased length of hospitalization, healthcare costs and increased mortality. Nonetheless, vancomycin 1 was considered as the standard therapy. However, the limitations of vancomycin 1 application in clinical practice were represented by a progressive increase in Methicillin Resistant S. aureus (MRSA) Minimum Inhibitory Concentrations (MIC), drug-related toxicity, and the lack of an oral formulation. Vancomycin 1 might be inferior to beta-lactams for bacteremia treatment because of Methicillin Susceptible Staphylococcus aureus (MSSA) in children. Vancomycin 1, a glycopeptide antibiotic, was first isolated in 1953 as a natural occurring antimicrobial agent produced by soil bacterium Amycolatopsis orientalis. Vancomycin 1 was approved for use in 1958 as a common antibiotic in treating rapidly growing penicillin-resistant Staphylococcus species. However, this antibiotic was not a primary choice of the treatment owing to its poor oral bioavailability and its various adverse effects. The evolution of Pseudomembranous enterocolitis, coupled with the spread of Methicillin Resistant Staphylococcus aureus (MRSA), led to the re-emergence of positive bacteria that were resistant to less-toxic agents. In addition, vancomycin 1 was used in patients who were intolerant or allergic to beta-lactams. This review encapsulates the strategies for the clinical management of MRSA.
Keywords
Hypofunctional, Occlusal, Root morphology, PDL space
Introduction
After the introduction of antibiotics, the evolution of antimicrobial resistance accelerated rabidly over the last decade which focused attention on the sustainability of recent modern medical practices globally. Staphylococcus aureus caused wide spectrum of pyogenic lesions involving several organs, hospital outbreaks and community acquired infections. In hospitals, S. aureus infections were lethal accompanied with resistance to several beta-lactam antibiotics [1]. Since 1970, MRSA (Methicillin Resistant Staphylococcus aureus), classified as pandemic pathogen, was concurrent with several nosocomial outbreaks and life threatening cross infections [2]. It was estimated that 2 billion populations globally might carry S. aureus on their skin and mucous membrane [3]. The over usage of antibiotics in hospitals was associated in the development of in-hospital acquired MRSA strains. Furthermore, increased use of antibiotics in animal feed has resulted in emergence of a new MRSA with multiple non-beta lactam drug resistance [4].
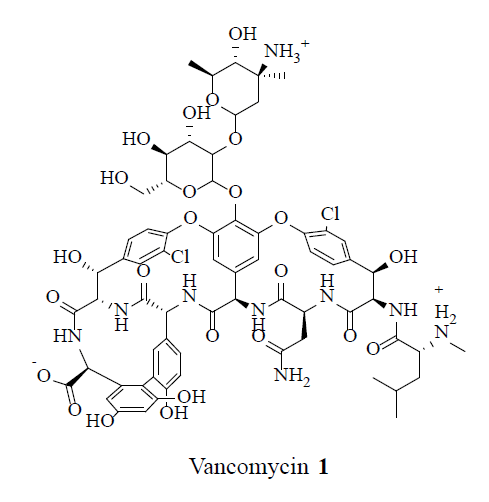
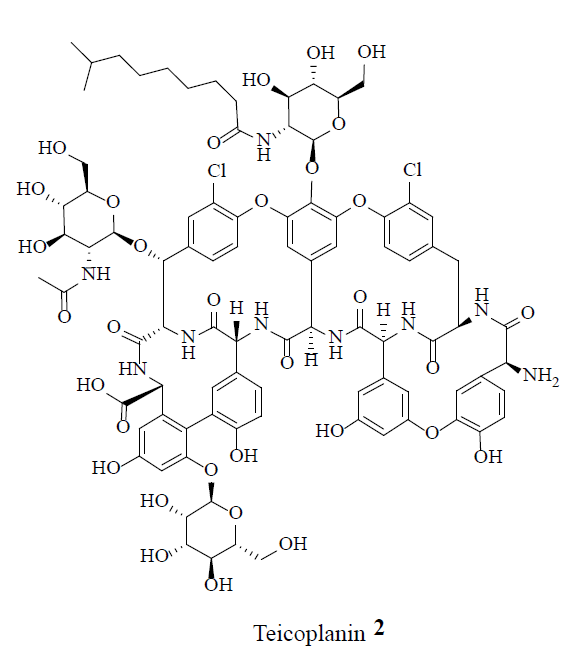
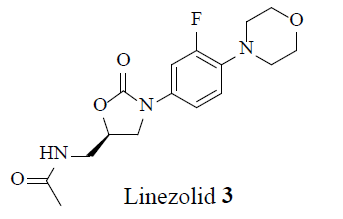
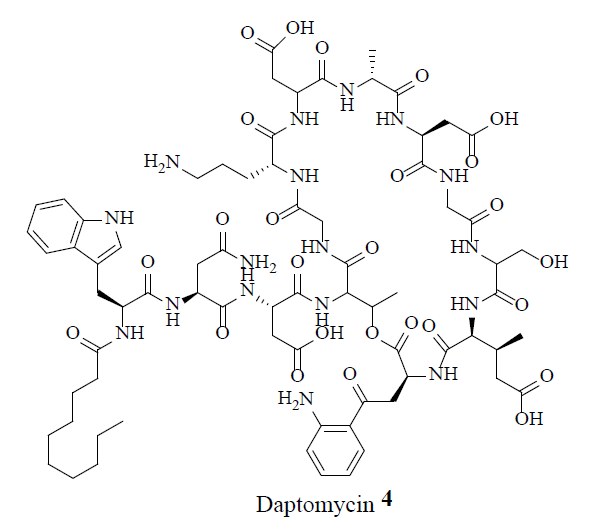
MRSA strains are highly variable in different geographical areas [5]. The transmission of Staphylococcal infections can be effectively curbed by restricting the over usage of antibiotics. Vancomycin (1), teicoplanin (2), linezolid (3) and daptomycin (4) were considered the conventional anti-MRSA drugs.
Nonetheless, throughout the world MRSA strains developed resistance to some of these drugs. Vancomycin (1) resistant and intermediate MRSA strains were reported intermittently. Recently, novel bioactive natural products had been identified with the anti-MRSA activity. These natural products have potential in the treatment of MRSA infections [6]. This review illustrates the anti-MRSA activities of conventional antibiotics and bioactive natural products.
Novel antimicrobial and resistance-modifying agents can be isolated from natural sources, such as land plants [7-9], fungi, lichens, endophytes, marine plants, seaweeds, corals, and other marine microorganisms. The development of novel genomic-type approaches in the search of small antibacterial molecules is an important avenue for the identification of future novel opportunities. The merit of natural products for the disruption of bacterial biofilms and their antibacterial role in foods was observed [10].
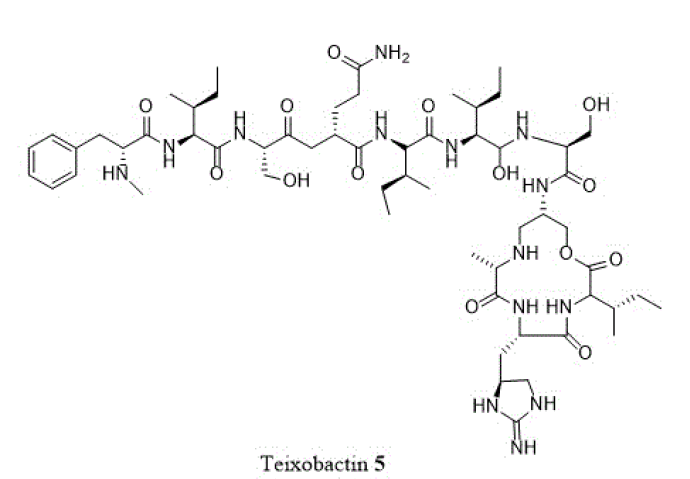
The single most important discovery in recent years has been that of the structurally unusual depsipeptide, teixobactin (5). This generated great interest for its intrinsic antibacterial activity, apart from its method of identification and culture of producing bacterium in its natural soil environment [11]. Since the original isolation was reported, new analogues were
synthesized and evaluated in a very short time [12]. Teixobactin (5) was observed to inhibit cell wall synthesis by binding at both lipids II and III with an activity against a wide panel of gram-positive organisms, endured potency in the presence of blood serum and exhibited microsomal stability with low toxicity [11]. Furthermore, teixobactin (5) was effective in vivo against S. aureus and Streptococcus pneumonia.
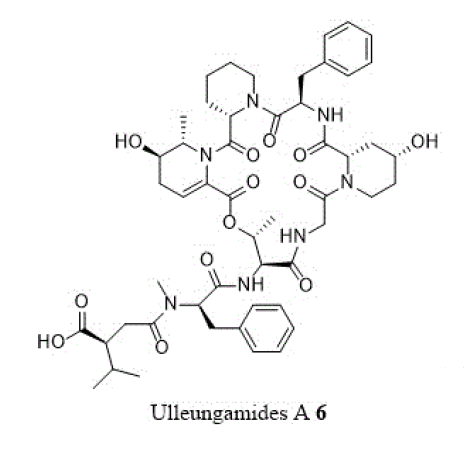
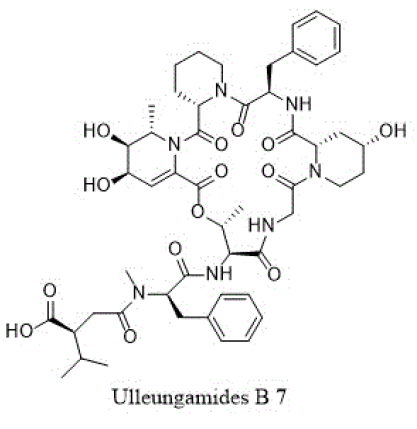
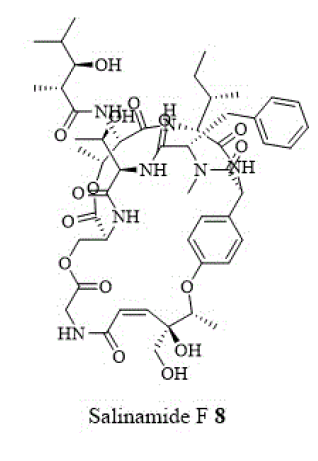
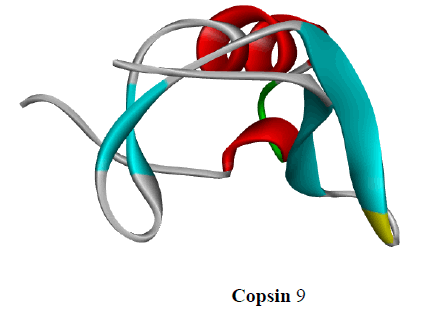
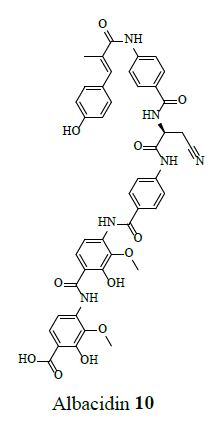
Other cyclic depsipeptides, ulleungamides A and B (6 and 7) exhibited inadequate antibacterial activity against S. aureus and Salmonella typhimurium with no cytotoxicity [13]. Salinamide F (8) showed significant activity against gram-positive and gram-negative bacterial RNA polymerase [10]. Copsin (9) an antibiotic peptide, isolated from a co-cultivated fungal source, stabilized by six cysteine bonds, exhibited good activity against gram-positive bacteria B. subtilis, Listeria spp., and Enterococcus spp., including vancomycin-resistant E. faecium. Furthermore, copsin (9) curbed cell wall biosynthesis through the inhibition at lipid II [10]. Albicidin (10) was a potent inhibitor of bacterial DNA gyrase (topoisomerase II) with excellent activity against a wide range of gram-positive and gram-negative bacteria. Cystobactamids were bacterial DNA gyrase (topoisomerase II) inhibitors and were effective against E. coli, A. baumannii, E. faecalis, S aureus, and S. pneumonia [14].
Antibiotic Resistance Mechanisms
The evolution of antibiotic resistance was assigned to a
combination of microbial characteristics. They were categorized as the selective pressure of antibiotic use and social and technical changes that increased the transmission of resistant organisms. The use as well as misuse of antibiotics in humans, nutritive and therapeutic antibiotic treatment of farm animals resulted in antibiotic-resistant bacteria. These mutant bacteria could enter into human food supply; colonize human digestive tract thereby transferring resistance genes to human commensals. Bacteria acquired the resistant genes from other bacteria when they joined together and transferred the genes to each other. Free plasmids were gathered by the bacteria which resulted in the resistance to a number of antibiotics. Once a resistant gene was localized in the bacterium DNA, the bacterium could dominate other bacteria and pass on the resistance gene to all of its descendants. The antibiotic resistance was initiated by certain mechanisms: A). Antibiotic inactivation-direct inactivation of the active antibiotic molecule; B). Target modification-alteration of the sensitivity to the antibiotic by target modification; C). Efflux pumps and outer membrane permeability changes-reduction of the drug concentration without modification of the compound; or D). Target bypass-some bacteria become refractory to specific antibiotics by bypassing the inactivation of a given enzyme. There was a great diversity of antibiotic resistance mechanisms within each of these four categories and a single bacterial strain. The diversity exhibited was due to several types of resistance mechanisms depending on the nature of antibiotics, their target site, and the bacterial species and whether it was mediated by a resistance plasmid or by a chromosomal mutation [1].
Drug Development for MRSA
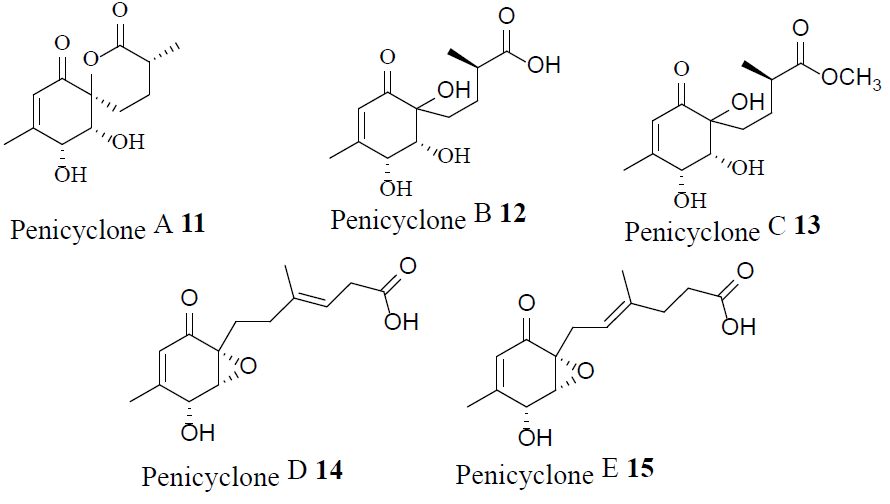
Penicyclones A-E (11-15)-antibacterial polyketides isolated from a deep-sea Penicillium species-showed potent antimicrobial activity against the gram-positive bacterium S. aureus, and were not cytotoxic [15].
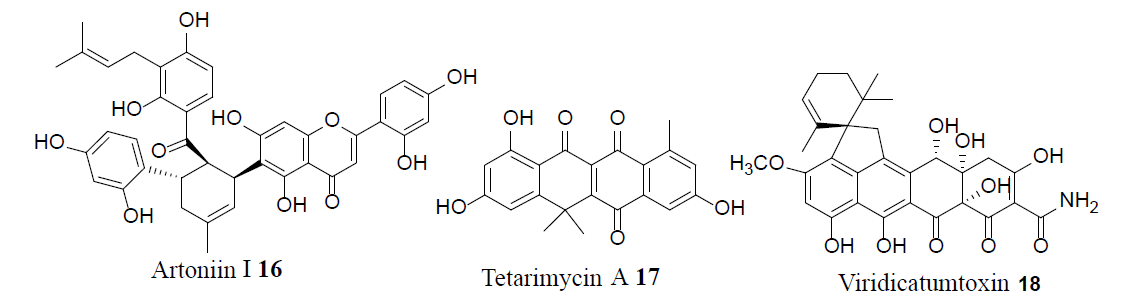
Combining existing antibiotics with the efflux pump inhibitor Artonin I (16) lowered MIC values against multidrug resistant S. aureus [16]. The synthetic methodology of the first total synthesis of tetarimycin A (17) was applied to generate new analogues, some of which were shown to be active against both methicillin-resistant S. aureus (MRSA) and Vancomycin- Resistant Enterococci (VRE). The presence of a crucial hydroxyl group within these analogues is vital for antibacterial activity [17]. Viridicatumtoxin (18), a structurally modified tetracycline antibiotic, exhibited excellent inhibitory activity against both MRSA and VRE. Furthermore, compound 18 presented slight cytotoxicity, while also exhibiting higher acid stability and bioactivity compared to the corresponding tetracycline antibiotics [18].
The synergistic activity of epigallocatechin gallate (19) and quercetin (20) when used in combination showed activity against a topical MRSA infection [19]. Heraclemycins A–D (21-24) were isolated from an endophytic Streptomyces species, where heraclemycin C (23) showing selective anti-tubercular activity; while heraclemycin D (24) was active against S. aureus, MRSA, and B. subtilis [20]. Totarol (25) produced methicillin-like activity, through the interference of PBP2’ expression, against MRSA [21].
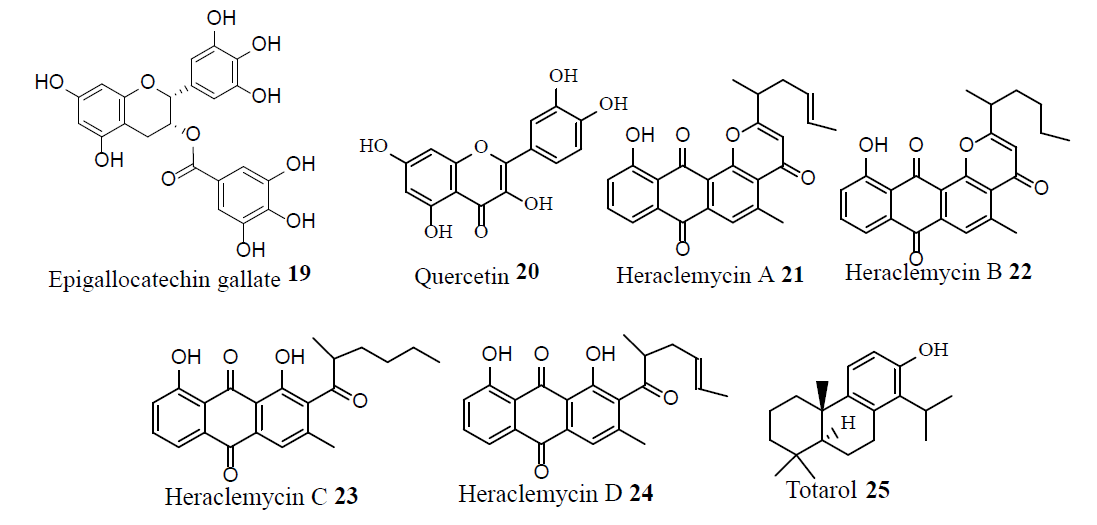
Tellimagrandin I (26), corilagin (27), and oxacillin (28) exhibited synergistic activity against MRSA strains by inhibiting the PBP2’ activity. Furthermore, compound 26 exhibited synergistic activity with tetracycline (29), against some MRSA strains [22].
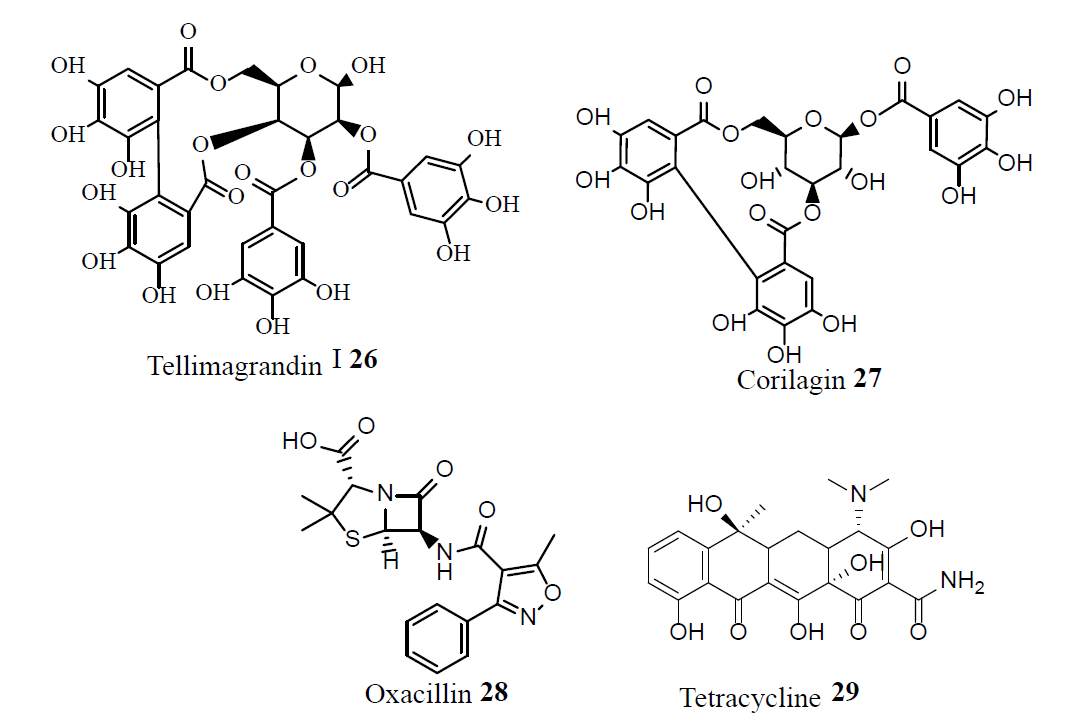
Guaianolide (30), a sesqiterpene lactone from Artemisia gilvescens, exhibited excellent activity against a clinical strain of MRSA [23]. Mansonone F (31) from Ulmus davidiana, showed exalted activity against MRSA strains [24]. Xanthorrhizol (32) an aromatic sesquiterpene phenol isolated from Curcuma xanthorrhiza; and compound 33, a stilbenoid-monoterpene isolated from Machaerium multiflorum; were active against MRSA. The alkyl esters of alkanin (34) exhibited activity against MRSA. Combinations of vancomycin (1), compound 35 and sophoraflavanone G (36) showed strong synergism against MRSA. Compound 37 and alopecurone B (38) showed activity against MRSA. Compared to (1), the phytochemicals Napthopyrone (39) and rubraxanthone (40) exhibited greater potency against MRSA [25].
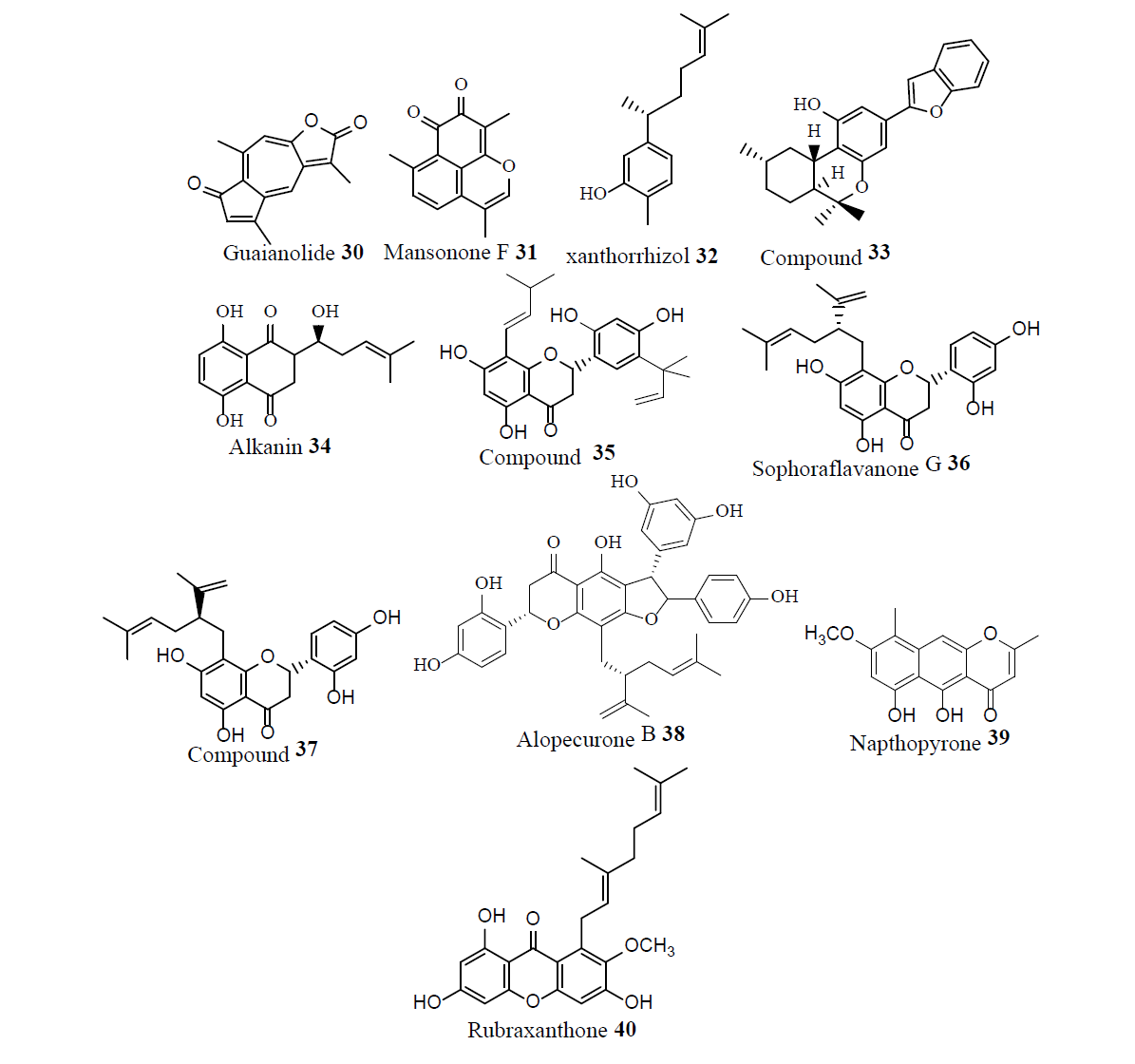
Following the emergence of MRSA, in 1958 vancomycin, 1, became the drug of choice to treat MRSA infections. Even after thirty years of use, vancomycin resistance was not observed, which lef to the conclusion that the development of vancomycin resistance might be impossible. However, in 1987 concern arose over the emergence of Vancomycin- Resistance S. aureus (VRSA) as a result of horizontal gene transfer from resistant Enterococci. In 2002, vancomycin resistance strain of MRSA suggested that the glycopeptide antibiotic treatment was not guaranteed. Novel intravenous drug combinations-comprised of Linezolid (Zyvox) 3, Synercid [a combination of quinupristin 41 and dalfopristin 42] and inert powder-were utilized to treat MRSA infections and combat this issue [26].
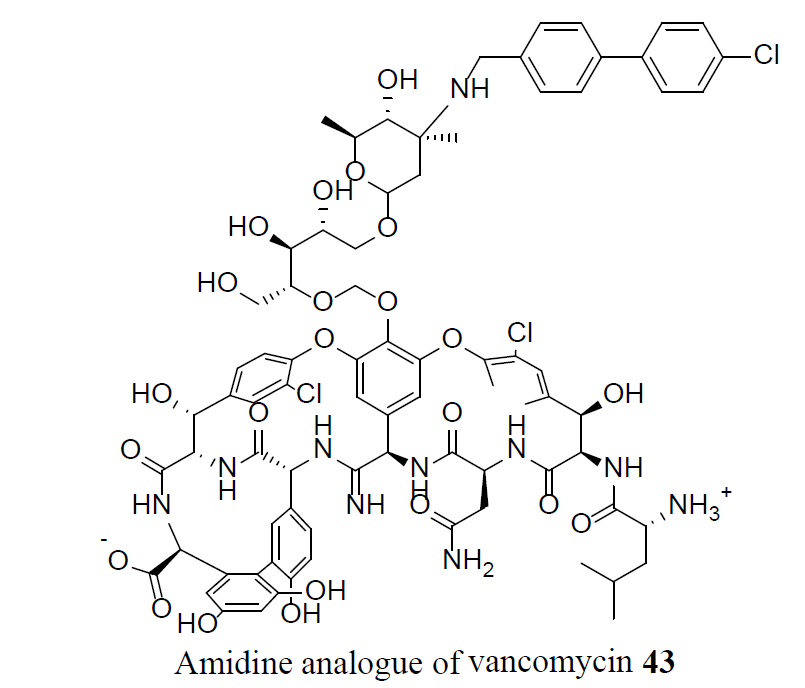
Vancomycin, 1, is lethal to bacteria by the disruption of bacterial cell-wall biosynthesis. The bacterium developed resistance by substituting an amino acid in a cell-wall component, preventing 1 from binding. A synthetic amidine analogue of vancomycin, 43, restored the binding to the modified cell wall component, which resulted in an activity against VRSA sensitive bacteria. Furthermore, compound 43 showed 10-100 times increased potency compared to vancomycin 1, against VRSA [27].
Conclusion
Over usage of antibiotics in agriculture and human medicine has resulted in global antimicrobial resistance epidemics. The danger of a MRSA explosion is rising in both the developed and developing countries. Bacteria have adopted diversified molecular mechanisms to acquire antibiotic resistance by the fine-tuning of genetic expression. In order to develop new approaches to overcome this problem, knowledge of the antibiotic resistance mechanisms is highly desirable. One strategy is to develop the inhibitors of resistance enzymes, administered as co-drugs with the antibiotics. Another strategy is to improve the delivery mechanisms of antibiotics to the sites of action to combat MRSA infections.
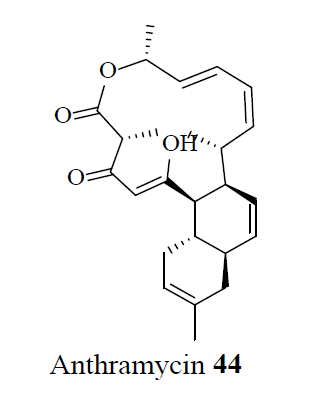
Plants produce natural products with diversified chemical structures that defend against microbes and their associated MDR mechanisms. Recently, anthracimycin 44, a new antibiotic drug developed from the marine microorganism, Streptomyces, was found to be effective against MRSA [28].
As such, the continuous discovery and development of compounds with novel antibiotic mechanisms of action is paramount in the fight against MRSA.
References
- Dzidic S, Susovic J, Kos B. “Antibiotic resistance mechanisms in bacteria: biochemical and genetic aspects.” Food Technol Biotechnol. 2008;46:11-21.
- Ayliffe GAG. “The progressive intercontinental spread of methicillin-resistant Staphylococcus aureus.” Clin Infect Dis. 1997;24:S74-9.
- Matouskova I, Janout V. “Current knowledge of methicillin-resistant Staphylococcus aureus and community-associated methillicin-resistance Staphylococcus aureus.” Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2008;152:191-202.
- Cockfield JD, Pathak S, Edgeworth JD, et al. “Rapid determination of hospital-acquired methicillin-resistant Staphylococcus aureus lineages.” J Med Microbiol. 2007;56:614-9.
- Shaffer RK. “The challenge of antibiotic-resistant Staphylococcus: lessons from hospital nurseries in the mid-20th century.” Yale J Biol Med. 2013;86:261-70.
- Hsieh YC, Lin YC, Huang YC. “Vancomycin, teicoplanin, daptomycin, and linezolid MIC creap in methhicillin-resistant Staphylococcus aureus is associated with clonality.” Mecicine (Baltimore). 2016;95:e5016.
- Singh RP, Kunari P, Reddy CR. “Antimicrobial compounds from sea-weeds-associated bacteria and fungi.” Appl Microbiol Biotechnol 2015;99:1571-86.
- Dobretsov S, Al-Wahaibi ASM, Lai D, et al. “Inhibition of bacterial fouling by soft coral natural products.” Int Biodeterior Biodegrad. 2015;98:53-8.
- El Amraoui B, El Amraoui M, Cohen N, et al. “Antifungal and antibacterial activity of marine microorganisms.” Ann Pharm Fr. 2014;72:107-11.
- Moloney MG. “Natural products as a source for novel antibiotics.” Trends in Pharmacol Sci. 2016;37:689-701.
- Ling LL, Schneider T, Peoples AJ, et al. “A new antibiotic kills pathogens without detectable resistance.” Nature. 2015;517:455-9.
- Jad YE, Acosta GA, Naicker T, et al. “Synthesis and biological evaluation of a teixobactin analogue.” Org Lett. 2015;17:6182-5.
- Son S, Ko SK, Jang M, et al. “Ulleungamides A and B, modified a,b-dehydropipecolic acid containing cyclic depsipeptides from Streptomyces sp. KCB13F003.” Org Lett. 2015;17:4046-9.
- Baumann S, Herrmann J, Raju R, et al. “Cystobactamids: myxobacterial topo-isomerase inhibitors exhibiting potent antibacterial activity.” Angew Chem Int Ed Engl. 2014;53:14605-9.
- Guo W, Zhang Z, Zhu T, et al. “Penicyclones A–E, antibacterial polyketides from the deep-sea-derived fungus Penicillium SP F23-2.” J Nat Prod. 2015;78:2699-703.
- Farooq S, Wahab AT, Fozing CD, et al. “Artonin I inhibits multidrug resistance in Staphylococcus aureus and potentiates the action of inactive antibiotics in vitro.” J Appl Microbiol. 2014;117:996-1011.
- Huang J, Lauderdale TY, Shia KS. “Studies on antibiotics active against resistant bacteria. Total synthesis of MRSA-active tetarimycin A and its analogues.” Org Lett. 2015;17:4248-51.
- Shang Z, Salim AA, Khalil Z, et al. “Viridicatumtoxins: Expanding on a rare tetracycline antibiotic scaffold." J Org Chem. 2015;80:12501-8.
- Betts JW, Sharill AS, Phee LM, et al. “In vitro activity of epigallocatechin gallate and quercetin alone and in combination versus clinical isolates of methicillin-resistant Staphylococcus aureus.” Org Lett. 2015;78: 2145-8.
- Liu M, Abdel-Mageed, Ren WM, et al. “Endophytic Streptomyces sp Y3111 from traditional Chinese medicine produced antitubercular pluramy-cins.” Appl Microbiol Biotechnol. 2014;98:1077-85.
- Nicolson K, Evans G, O’toole PW. “Potentiation of methicillin activity against methicillin-resistant Staphylococcus aureus by diterpenes.” FEMS Microbiol Lett. 1999;179:233-9.
- Shiota S, Shimizu M, Mizusima T, et al. “Restoration of effectiveness of b-lactams on methicillin-resistant Staphylococcus aureus by tellimagrandin I from rose red.” FEMS Microbiol Lett. 2000;185:135-8.
- Sato Y, Oketani H, Yamada T, et al. “A xanthanolide with potent antibacterial activity against methicillin-resistant Staphylococcus aureus.” J Pharm Pharmacol. 1997;49:1042-4.
- Shin D, Kim H, Min K, et al. “Isolation of a potent anti-MRSA sesquiterpenoid quinone from Ulmus davidiana var. japonica.” Chem Pharm Bull (Tokyo). 2000;48:1805-6.
- Gibbons S. “Anti-Staphylococcal plant natural products.” Nat Prod Rep. 2004;21:263-77.
- Tsiodras S, Gold HS, Sakoulas G, et al. “Linezolid resistance in a clinical isolate of Staphylococcus aureus.” Lancet. 2001;358:207.
- James RC, Pierce JG, Okano A, et al. “Redesign of glycopeptide antibiotic-back to the future.” ACS Chem Biol. 2012;7(5):797-804.
- Hensler ME, Jang KH, Thienphrapa W, et al. “Anthracimycin activity against contemporary methicillin-resistant Staphylococcus aureus.” J Antibiotics. 2014;67:549-53.