Research Article - Journal of Parasitic Diseases: Diagnosis and Therapy (2023) Volume 8, Issue 2
Genotyping and phylogenic relationship of giardia isolates from Sudanese suspected patients using (TPI) triose phosphate isomerase gene.
Anas M Elnazeer1*, Samah Hassan Suliaman2, Mubarak Mustafa3, Ishraga Adam Elzain4, Shafag Hassan Kirkusawi51The International University of Africa, Faculty of Pure and applied sciences department of Microbiology, supervisor of the molecular biology research lab, Sudan
2Faculty of Medicine, Al Zaiem Al Azhari University, Sudan
3The International University of Africa, faculty of medical laboratory sciences, department of Parasitology, Sudan
4Tropical Medicine Research Institute, National Research Centre, Sudan, Khartoum, Sudan
5The International University of Africa, faculty of agricultural production and processing technology, Sudan
- *Corresponding Author:
- Anas M Elnazeer
The International University of Africa
Faculty of Pure and applied sciences department of Microbiology
supervisor of the molecular biology research lab, Sudan
E-mail: anas-sci@iua.edu.sd
Received: 19-Mar-2023, Manuscript No. AAPDDT-23-92238; Editor assigned: 20-Mar-2023, Pre QC No. AAPDDT-23-92238(PQ); Reviewed: 24-Mar-2023, QC No. AAPDDT-23-92238; Revised: 24-Mar-2023, Manuscript No. AAPDDT-23-92238(R); Published: 31-Mar-2023, DOI: 10.35841/2591-7846-8.2.136
Citation: Elnazeer AM, Suliaman SH, Mustafa M, et al. Genotyping and phylogenic relationship of giardia isolates from Sudanese suspected patients using (Tpi) triose phosphate isomerase gene. J Parasit Dis Diagn Ther 2023;8(2):136
Abstract
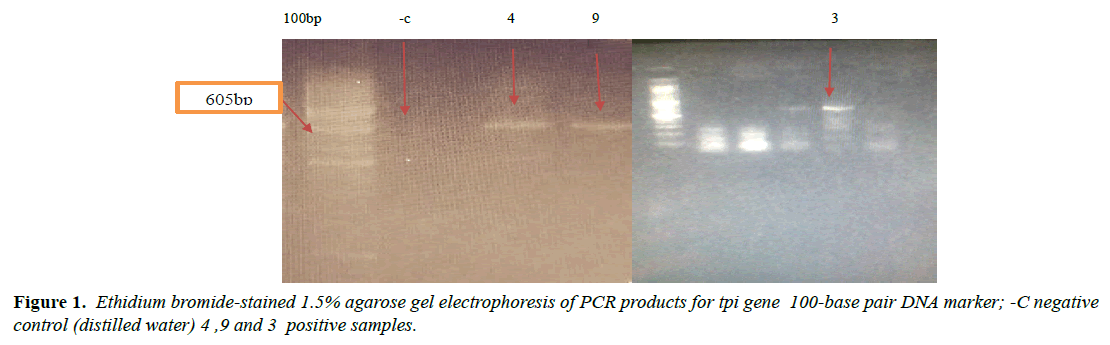
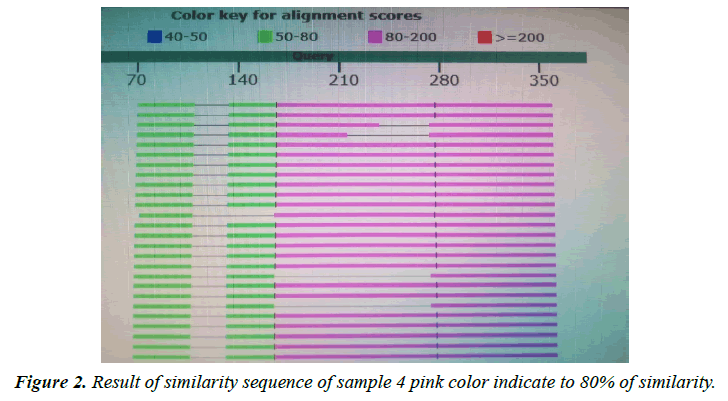
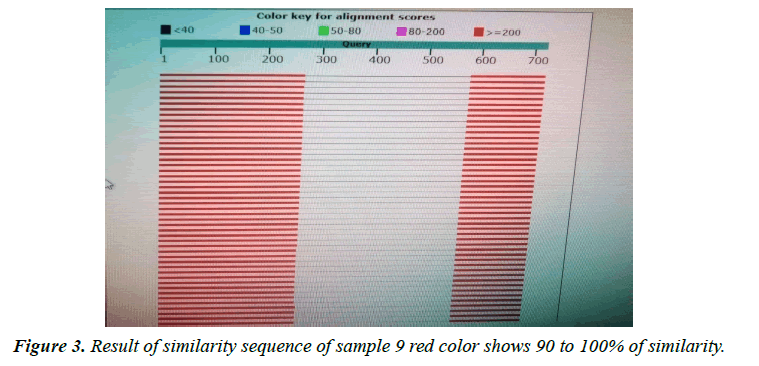
Giardia lamblia is one of the most common intestinal pathogens in both humans and animals throughout the world, globally, there are greater than 700,000 deaths per year associated with diarrheal disease, and understanding the organism requires more in-depth knowledge of its biology. The current study amid to establish a research platform for the molecular characterization of Giardia isolates in Sudan, focusing on the biology of giardia, through, molecular characterization attempts to genotyping A laboratory-based experimental study was designated. A total of 30 fecal samples of giardia positive from suspected patients after informed consent were used in this study from three states. Molecular characterization of giardia isolates which included DNA extraction by Guanidine HCL after liquid nitrogen treatment. PCR detection and sequencing were obtained. PCR results showed that only 5 samples were positive out of 30 that represent 17% total of cyst samples when using tpi gene 605 bp, in the Sequencing results only two samples were successfully sequenced (one from Khartoum and one from Sinnar states) while bioinformatics results showed similarity between this isolates through alignment and multiple alignments to giardia in Africa. After sequencing of tpi gene, the Sudanese isolates from Khartoum state were found to be identical to genotype (A) with 80%, while samples from Sinnar State were identical to genotype (B) with 90%.
Keywords
Genotyping, Tpi gene, Alignment, Sinnar, Giardia.
Introduction
Giardia was one of the first protozoans to be described. In1681 van Leeuwenhoek discovered the trophozoites of this genus. The Dutch microscopist made glass lenses and set them into metal frames which he made into simple microscopes. Globally, there are greater than 700,000 deaths per year associated with diarrheal disease. The flagellated intestinal parasite, giardia lamblia is one of the most common intestinal pathogens in both humans and animals throughout the world [1] G. intestinalis has a wide genetic variety and it is characterization helps in the understanding of it is transmission dynamic [2] PCR genotypes classification of Giardia in feces depends on quantity and quality of purified DNA and the removal of a great number of inhibitors [3] Currently, molecular genotyping of G. intestinalis is based on analyses of one or several of the following genetic loci: small subunit ribosomal DNA, elongation factor 1-α, histone 2b, and histone 4, β-giardin, glutamate dehydrogenase, and triose phosphate isomerase (ssrDNA, ef, h2b, h4, bg, gdh and tpi respectively). The first four (ssrDNA, ef, h2b, h4) are considered conserved genetic markers and the latter three (bg, gdh, tpi) are considered more variable [4]. Since this work mainly focuses on G. intestinalis infection in humans only genotyping of the two human infecting assemblages A and B is described. Meta-analyses of several molecular typing studies of the human infecting assemblages A and B have shown that assemblage B Giardia is more commonly occurring than assemblage A, in humans [5,6]. The currently utilized loci and especially the more variable loci, provide ample discrimination of assemblage B isolates, where as little discrimination is found among assemblage A isolates [4-6]. The human infecting G. intestinalis assemblages (A and B) have been divided into different sub-assemblages based on sequencing results from the bg, gdh and tpi loci. Assemblage A is grouped into AI, AII, and AIII, where AII can be further subdivided intoAII-1 and AII-2 based on two nucleotide substitutions on the bg locus. AII is described to typically infect humans, AI has been found in humans but mainly infects animals and AIII has exclusively been found in animals [5-7]. The routinely used markers for sequence based genotyping of assemblage A provide low resolution between different A isolates, and there is a high necessity to find new molecular markers for genotyping of assemblage A G. intestinalis. Sequence-based genotyping of assemblage B has been hampered due to a high frequency of mixed base polymorphisms, seen as double peaks at single nucleotide positions in the sequencing chromatograms [6,7]. Although there is a high level of difference in the sequences between different B isolates, there is currently no functional way of categorizing them into different sub-assemblages this is due the high frequency of mixed base polymorphisms found in the majority of sequences from clinical samples.
Materials and Methods
Sources of DNA
A total of 30 (10 khartoum,10 Sinnar, 10 Gezira) fecal samples were used for DNA extraction ,sample size was calculated by the formula suggested by [8] N ≥ 50+8p where p is number of predictors if we predictor (p) =1 in addition to other samples from culture used as control positive ,fecal samples were purified and washed in phosphate buffer saline PBS then confirmed microscopically and counted before used in DNA extraction and preserved -20ºc Ethical approval for the current study was obtained from ethical committee of Tropical Medicine Research Institute (TMRI) Sudan, Khartoum . Standard equipment used in a microbiology and molecular biology laboratory was used.
DNA extraction method
DNA extraction was conducted According to a protocol designated by [9-12] with slightly modification to optimize disruption of the cysts prior to DNA extraction Cyst samples were treated by boiling in 100ºC for 2 min and put in liquid nitrogen for 2min these steps were repeated 3times before DNA extracting a guanidine hydrochloride method was used as the following.
1 ml of sample added to 1ml lyses buffer 2% SDS, 1ml guanidine hydrochloride, 300μl ammonium acetate ,and 10μl proteinase K and incubated at 37°C overnight , DNA was harvested, then confirmed and evaluated by scanning Nano drop system for reading DNA and protein concentration, and kept at -20°C as stock for further PCR experiments.
PCR performed According to [13] with slightly modification Using Triose phosphate isomerase gene TPI fragment 605 bp was amplified by using primers AL3543 [5-AAATIATGCCTGCTCGTCG-3] forward and AL3546 [5-CAAACCTTITCCGCAAACC-3] reverse as target gene sequence. The reactions were performed by using redy prepared primex (MiQ COMPANY) 20 μl commercial product contain (200 μmd NTP, 1XPCR Buffer,3mM MgCl2,5U of Tag polymerase). In the reaction we add 1.5 μl forwards primer and 1.5 μl reverse one 10μl DNA template and complete the volume to 20μlwith sterile D.W. and the reaction was conducted at 94ºc for 5min an initial temperature 94ºc for 30 sec, 55ºc for 45 sec, 72ºc for 2 min, 72ºc for 10 min to complete 30 cycle a PCR product was confirmed by run in 1.5% agarose with ethidium promide stain 1% read by Gel Documentation System for target visible bands then record the result. PCR from feces depend on the quality and quantity of purified DNA and removal of great variety of inhibitors such as biliry salts, bilirubin, and 10% of formalin can inhibit PCR [3]. This test used in order to control PCR inhibitors substances which may found in faecal materials. The test conducted by added 10μl DNA template which give positive PCR to that give negative and run it in thermo cycler with same PCR program and the results were records.
Bioinformatics, sequencing and phylogeny analysis
Sequencing was done by www.Bioneer.com Korea to four PCR positive samples Alignment for sequences was obtained by Basic Local Alignment Search Tool (BLAST) www.ncbi. com which allows rapid sequence comparison of a query sequence against a database in order to compare the similarity between the sequences and other reference sequences on DATA base, DNA sequences are homologous if they are at least 70% identical, and there are three steps to run BLAST. (1) Choose the sequence (query) (2) Select the BLAST program (3) Choose the database to search Then click BLAST Aphylo genetic tree and multiples alignment was conducted by (clusterW and cluster Omega) software programs in order to study the relationships between different isolates and to compare with other sequences from Data base.
Results
The variant in similarity sequences were subjected to multiple alignment, using ClusterW and cluster Omega software, matching with tpi nucleotide sequences of Giardia intestinalis assemblage A and B isolate deposited from gene bank under ID (KY444791.1), (gi|1255657882|gb|KY444788.1). ( g i | 1 2 5 5 6 5 7 8 7 9 | g b | K Y 4 4 4 7 8 7 . 1 ) , (gi|340885174|gb|HQ836660.1) (LN626349), (LN626350.1). Result in (Figures 1-4) pair alignment between two sequences sample1 and sample2 shows identity loci ( marked with stars ⃰ ⃰ ⃰ ⃰ and gabs marked ------) there was identic between both samples. (Figure 5) Multiple alignment sequences between different Giardia isolates and sample1,2 showing identity and gabs (Figure 6) Multiple alignment sequences between different Giardia isolates and sample1,2 showing identity and gabs .The result show that sample1 identity 80% to genotype (A) or assemblage A, while sample2 identity 90%to genotype (B) assemblage B.
Phylogeny relationships obtained by using ClusterW and cluster Omega software in order to study the relationships between different isolates. (Figure 7) a phylogram which show that sample 2 significantly related to assemblage B (KY444791.1) both were branched from one root assemblage B. (Figure 8) a phylogram which show that sample 1 significantly related to assemblage A .(gi|1255657879|gb|KY444787.1) both were branched from one root assemblage A. (Figure 9). A phylogram which show that both samples 1,2 were related to each other. While sample 4 was close related to assemblage A and sample 9 related to B.
Discussion
Molecular characterization was studied; DNA extraction was conducted by Guanidine HCL after Liquid nitrogen treatment. PCR result show that only 5 samples were positive out of 30 that represent 17% from total of cyst samples (Table 1) (Figure1) when using tpi gene 605bp lower grade of PCR may due to DNA conc, purity and PCR inhibitors, but it considered high sensitive. Sequencing result obtained by bioneer company(Korea) only two samples were successfully sequenced (sample 1 Khartoum and 2 Sinnar), bioinformatics results similarity in alignment and multiple alignment show that sample1 identity 80% to genotype (A) or assemblage A, while sample2 identity 90%to genotype (B) assemblage B (Figures 4-6). Phylogeny relationships obtain by using ClusterW and cluster Omega software in order to study the relationships between different isolate (Figures 7-9) which show that sample 9 significantly related to assemblage B (KY444791.1) while sample4 related to assemblage A.(gi|1255657879|gb|KY444787.1) and both samples 4,9 are each related to other (Figure 9) Similar result was obtained by [13] in Pakistan [14] confirmed PCR as sensitive technique in compared with ELISA and microscopic technique when used gdh gene which obtain that PCR more specific and sensitive than other techniques , In addition, the genetic characterization of the parasite was carried out by amplifying and sequencing the glutamate dehydrogenase (gdh) gene in southern Brazil [15], the DNA sequences revealed that 7 (36.8%) out of 19 isolates belonged to assemblage B, while 6 of them (31.6%) belonged to assemblage C, 5 (26.3%) to assemblage A and 1 (5.3%) to assemblage D this is other supportive study to our finding which concluded that the giardia isolates from Sudanese suspected patients was found resemble to geno type B after sequencing of tpi gene and bioinformatics analysis.
| Locality | PCR +VE | PCR –VE | TOTAL |
|---|---|---|---|
| Khartoum | 2(20%) | 8(80%) | 10 |
| Sinnar | 3(30%) | 7(70%) | 10 |
| Gezira | 0(0%) | 10(100%) | 10 |
| Total | 5(17%) | 25(83%) | 30 |
Table 1. PCR results.
References
- Fisher BS, Estraño CE, Cole JA. Modeling long-term host cell-Giardia lamblia interactions in an in vitro co-culture system. PloS one. 2013;8(12):e81104.
- Nunes BC, Calegar DA, Pavan MG, et al. Genetic diversity of Giardia duodenalis circulating in three Brazilian biomes. Inf Gen and Evolution. 2018;59:107-12.
- Nora M, Daniela P, Marta M, et al. PCR amplification of triose phosphate isomerase gene of Giardia lamblia in formalin fixed feces, Rev Latinoam Microbial 2007;49:(1-2), 6-11.
- Wielinga C, Ryan U, Thompson RA, et al. Multi-locus analysis of Giardia duodenalis intra-Assemblage B substitution patterns in cloned culture isolates suggests sub-Assemblage B analyses will require multi-locus genotyping with conserved and variable genes. Int J Parasitol. 2011;41(5):495-503.
- Lebbad M, Mattsson JG, Christensson B, et al. From mouse to moose: multilocus genotyping of Giardia isolates from various animal species. Vet Parasitol 2010;168: 231-239
- Lebbad M, Petersson I, Karlsson L, et al. Multilocus genotyping of human Giardia isolates suggests limited zoonotic transmission and association between assemblage B and flatulence in children. PLoS Neglected Tropical Dis. 2011;5(8):e1262.
- Cacciò SM, Ryan U. Molecular epidemiology of giardiasis. Mol biochem parasitol. 2008;160(2):75-80.
- Green SB. How many subjects does it take to do a regression analysis. Multivar Behav Res 1991;26(3):499-510.
- Adamska M, Leońska-Duniec A, Maciejewska A, et al. Comparison of efficiency of various DNA extraction methods from cysts of Giardia intestinalis measured by PCR and TaqMan real time PCR. Parasite. 2010;17(4):299-305.
- David EB, Coradi ST, Oliveira-Sequeira TC, et al. Diagnosis of Giardia infections by PCR-based methods in children of an endemic area. J. Venom Anim Toxins Incl Trop Dis 2011;17:209-15.
- Elwin K, Fairclough HV, Hadfield SJ, et al. Giardia duodenalis typing from stools: a comparison of three approaches to extracting DNA, and validation of a probe-based real-time PCR typing assay. J Med Microbiol. 2014;63(1):38-44.
- Kasaei R, Carmena D, Jelowdar A, et al. Molecular genotyping of Giardia duodenalis in children from Behbahan, southwestern Iran. Parasitol Res 2018;117:1425-31.
- Sulaiman IM, Fayer R, Bern C, et al. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerging Infect Dis 2003;9(11):1444.
- Shakir AI. Molecular Characterization and Serological Cut-off (Copro-Antigen ELISA) of Entamoeba Histolytica and Giardia Lamblia-Khartoum State (Doctoral dissertation, Sudan University of Science & Technology).
- Jeske ST, Macedo MR, Bianchi T, et al. Molecular characterization of Giardia lamblia and risk factors for giardiasis among immunocompromised patients in southern Brazil. Braz J Bio 2022;9;82.
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref