Research Article - Biomedical Research (2017) Volume 28, Issue 11
Genetic diversity of the Striped Hamster is density- and seasonal-dependent
Laixiang Xu*, Huiliang Xue, Jinhui Xu and Lei ChenCollege of Life Sciences, Qufu Normal University, Qufu, Shandong, PR China
Accepted date: March 29, 2017
Abstract
Aim: The Striped Hamster (Cricetulus barabensis) is characterized by high reproductive ability, seasonal breeding feature, and fluctuation on population abundance in different years and seasons. Fluctuation on population abundance is assumed to affect genetic diversity. However, the relationship between the genetic diversity and the abundance of Striped Hamster population is less understood.
Methods: In this study, the Striped Hamsters were captured from Wu village of Shandong province in the autumn and spring from 2004-2008, and that both seasonal populations and yearly populations were studied. Genetic structure of the Striped Hamsters was studied by using DNA microsatellite markers. The relative density of the Striped Hamster was estimated by the rate of trap success. Temporal variation of the genetic structure and the trap success were analysed to determine the correlation between the genetic structure and the trap success.
Result: The results showed that the genetic structure and the population density were significantly fluctuated with the time. The genetic diversity of the Striped Hamster population was positively correlated with the population density and the increase of the genetic diversity was correlated with the decrease in population density between spring and autumn population.
Conclusion: The specific reasons for these results may be caused by density- and seasonal-dependent dispersal of the Striped Hamster, which is in accordance with Elton hypothesis.
Keywords
Striped Hamster, Genetic diversity, Population density, Season.
Introduction
The striped hamster is a dominant rodent species in northern China plain and is also distributed in Russia, Mongolia and Korea. The climate of the northern China is arid and characterized by warm and dry summer and cold winter with the minimum temperatures of -20°C in history [1,2]. The striped hamsters feed on stems and leaves of plant during summer and on foraging crop seeds in winter. They are solitary and very aggressive [3]. The lifetime of the hamsters is about 1.5 to 2.5 year. The reproductive cycle is about 2 months.
Many factors affect animal population fluctuation, and they can be divided into environmental and biotic factors. Environmental factors, such as temperature, precipitation, irrigation and others, act directly on the structure of the animal population, modify the spatial and temporal distribution of the species [4]. Biotic factors, including predation, competition, food availability and quality could make certain pressure on the population survival [5-7].
Seasonal change determines the critical environmental conditions. It is a major factor which influences population abundance. Heavy rainfall in summer causes a sharp reduction in the abundance of the hamsters and adequate food in autumn lead to an increase of the hamsters [1,7]. So, the abundance of the Striped Hamster populations fluctuates among seasons, and the effect of population abundance on genetic diversity among seasons needs to be investigated.
Dispersal movements have been identified as one of the most general causes of population synchrony [8]. The tundra vole exhibits negative density dependent dispersal. For example, both scarcity of mates and inbreeding avoidance may result in enhanced dispersal at low population density. Negative density-dependent dispersal is prevalent in voles while positive density-dependent dispersal is proposed in rodents [9,10]. Understanding the population fluctuation mechanism is very important to control the population abundance and maintain the ecological balance in ecology field [11].
Dramatic fluctuations in population abundance over time and space exist in many species, including small rodents [12], it may cause variations in genetic diversity of these populations [13]. The effect of the population abundance on the genetic diversity of the Striped Hamster is less understood.
Elton found fluctuation could reduce the population genetic diversity [14]. Elton hypothesised that fluctuations reduced population genetic variation and influenced the direction of selection pressures. Modern genetic tools make it possible to detect the genetic variation of the cycle populations and identify the adaptive genomic regions relevant to ecological factors [15,16].
However, the results of Xie and Zhang showed that the fluctuate population of the greater long-tailed hamsters lead to a high level of genetic diversity [17]. However, several researches have confirmed that fluctuated population could affect the genetic diversity. For example, the genetic variation in northern cyclic populations is higher [16]. And the populations of grey-sided (Myodes rufocanus) and red (Myodes rutilus) voles from northern Norway with abundance fluctuation characteristics also display higher genetic diversity [18]. Besides, the genetic variation of animal populations is fluctuated with population oscillation [19]. Generally, the genetic diversity increases with the population abundance [20]. But in some populations, the genetic diversity shifts in the opposite direction with the population abundance [21,22]. In conclusion, in different species, the correlation between the population abundance and the genetic diversity are variable.
Density-dependent dispersal is a main factor affecting the genetic diversity in northern cyclic populations [14]. But the population density has opposite dispersal effect among different species. For example, it has negative dispersal effect in arvicoline rodent [23,24], but has positive effect in water vole [20]. In the arvicoline rodent populations, the genetic diversity is maintained at a stable level throughout the cycle [25]. However, in some populations, genetic diversity can be affected by fluctuation and appear temporal oscillation [26]. Different density-dependent dispersal forms may lead to various effect on genetic diversity of the fluctuate populations.
The Striped Hamster is one kind of dominant agricultural rodent species in croplands of the North China Plain. Its population density is fluctuated over time [27]. Genetic diversity is very important for the animals to adapt the changing environment [28]. However, some environmental factors and biotic factors could affect the degree of the genetic diversity. But the correlation of the Striped Hamster population between the population abundance and the genetic diversity is still less understood. In this study, we mainly study the genetic variation of the Striped Hamster population from Wu village, Qufu County, Shandong province of the North China during 2004-2008.
Materials and Methods
Ethics statement
All the operation of handling the Striped Hamsters in this study were inspected and approved by the Ethics Committee of Qufu Normal University. All the researchers had received professional training and had the permission to conduct animal studies.
Samplings collection
Striped Hamsters were captured monthly during 2004-2008 using iron traps in Wu Village, Qufu city, Shandong province, China. The place is located in the (116°98’E, 35°58’N) North China Plain, and it has four seasons and a typically warm-temperate climate. The Striped Hamsters were live trapped using a brand of trap (Table S1).
The captured hamsters were euthanized by CO2 asphyxiation immediately, and given a unique number. Record the sex and weight, then dissect and measure the hamsters to provide estimates of body size, age, and reproductive condition. There are more than 20 permanent plots used as hamster trapping in Wu village. Each plot contained three parallel trapping lines with about 30 m apart from each neighbor trapping lines. About thirty traps were placed along every trapping line with an interval of 5 m. Trappings were carried out for 3 consecutive days in mid-month during 2004-2008 to avoid trapping too many animals. Population densities were defined as the percentage of the trap success (T%). Trap success was used as an index for the density of the Striped Hamsters. It is calculated as: T%=Total number of captured hamsters/total number of traps × 100%.
The populations in this study include spring, autumn, and yearly populations. The individuals of the spring populations were captured in spring (March, April and May), and those of the autumn populations were captured in autumn (September, October and November). The yearly populations are the sum of the individuals from spring and autumn populations. Because the amount of individuals captured in the winter (December, January and February )and summer (June, July and August) is too small, the genetic diversity of the winter and summer populations were not researched.
DNA extraction and genetic analyses
The genomic DNA was extracted from liver tissues using the traditional phenol-chloroform extraction method. Liver tissue is an ideal material for high DNA extraction efficiency for higher DNA content and no fat matrix exist in liver tissue than others. Nine microsatellite loci of the Striped Hamsters (Table S1) were used on the basis of its high amplification efficiency and rich polymorphism. The sequence of each locus was amplified by PCR including 50 mM KCl, 10 mM Tris-HCl, 2.5 mM MgCl2, 0.2 mM each dNTP, 1 U of Taq DNA polymerase (Promega), 10 pM forward and reverse primers, and approximately 2 ng of genomic DNA in total of 25 μL. The PCR procedures began with a 5 min pre-denaturing step at 94°C, followed by 30-35 cycles of the following reaction steps: denaturing at 94°C for 45 s, annealing at 46~58°C for 45 s, and extending at 72°C for 1 min, with a final extension step for 5 min at 72°C. All products were analysed in an ABI 377 instrument (Perkin-Elmer Applied Biosystems, Foster City, California) and the gel analysis was conducted by using GENESCAN3.1 (Perkin-Elmer Applied Biosystem).
Measures of genetic variation
Population genetic diversity was estimated over all loci within each population by the effective mean number of alleles per locus (Ne) [29], Polymorphism Information Content (PIC) [30], unbiased estimates of expected Heterozygosities (He) [31], and observed Heterozygosities (Ho) [32].
Statistical analysis
SPSS 15.0 software (SPSS Inc., Chicago, IL, USA) was used to analyse the data.
Results
The genetic diversity of the yearly populations
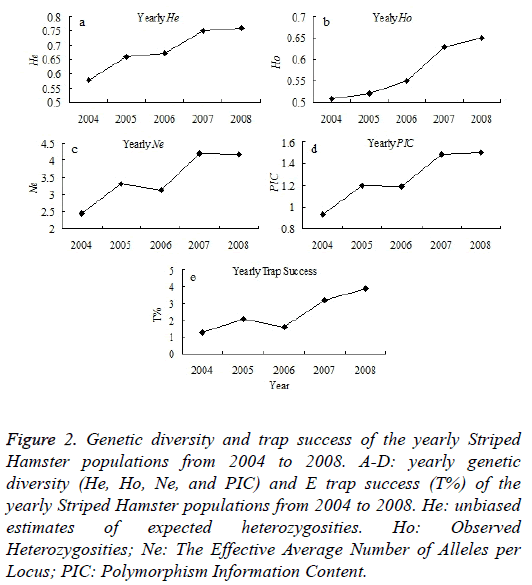
In this study, a total number of 298 Striped Hamsters were captured monthly during 2004-2008 using iron traps in Wu Village, Qufu city, Shandong province, China (Figure 1). The Striped Hamster populations present an obvious fluctuation during 2001-2013. In this study, the genetic diversities of the Striped Hamster populations from the Wu Village during 2004-2008 were examined. It shows that the genetic structure indicators of He, Ho, Ne and PIC appears an increase tendency, except for the Ne and PIC have little decrease in 2006 than 2005 (Figures 2A-2D). The population abundance shows an increase tendency during 2004-2008 (Figure 2E). The variation trend of the population abundance is accordance with that of the genetic diversity.
Figure 2: Genetic diversity and trap success of the yearly Striped Hamster populations from 2004 to 2008. A-D: yearly genetic diversity (He, Ho, Ne, and PIC) and E trap success (T%) of the yearly Striped Hamster populations from 2004 to 2008. He: unbiased estimates of expected heterozygosities. Ho: Observed Heterozygosities; Ne: The Effective Average Number of Alleles per Locus; PIC: Polymorphism Information Content.
The genetic diversity of the spring and autumn populations
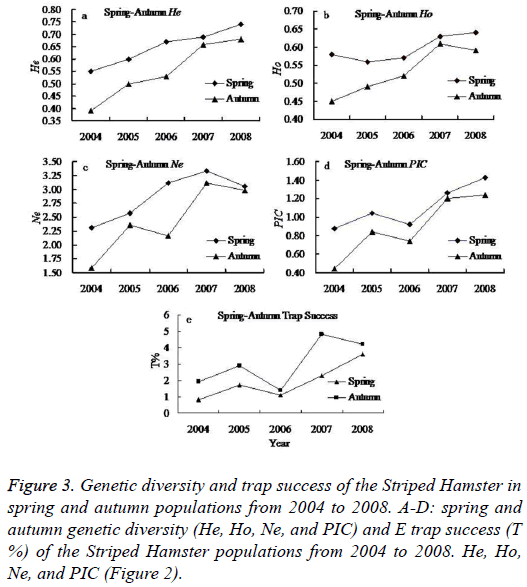
Seasonal change has important impact on the animal living environments (food availability and living temperature). It is a major factor influencing population abundance. In this study, we found higher population abundance in the spring and autumn than that in summer and winter. In addition, the autumn Striped Hamster population abundance was higher than that of the spring populations, but reverse magnitude relation were detected in genetic diversity between the spring and autumn populations (Figure 3).
Relationship between the population density and the genetic diversity
In this study, positive and significant correlation (P<0.05) was found between the genetic structure indicators of He, Ho, Ne and PIC and the trap success for the spring, autumn and the yearly populations of the Striped Hamster (Table 1), and the level between the genetic diversity and the trap success was highly significant (Table 1).
| Spring trap success | Autumn trap success | Yearly trap success | ||
|---|---|---|---|---|
| Spring Ne | r | 0.953393 | ||
| p | 0.035701* | |||
| Autumn Ne | r | 0.82614 | ||
| p | 0.0362673* | |||
| Yearly Ne | r | 0.944958 | ||
| p | 0.015796* | |||
| Spring PIC | r | 0.905377 | ||
| p | 0.034637* | |||
| Autumn PIC | r | 0.896414 | ||
| p | 0.039453* | |||
| Yearly PIC | r | 0.934681 | ||
| p | 0.019831* | |||
| Spring Ho | r | 0.896369 | ||
| p | 0.037231* | |||
| Autumn Ho | r | 0.933771 | ||
| p | 0.039479* | |||
| Yearly Ho | r | 0.954649 | ||
| p | 0.009584** | |||
| Spring He | r | 0.926385 | ||
| p | 0.031424* | |||
| Autumn He | r | 0.931758 | ||
| p | 0.0283567* | |||
| Yearly He | r | 0.920344 | ||
| p | 0.026728* | |||
Table 1: Pearson correlations between genetic diversity (Ne, PIC, He, and Ho) and trap success (T%) of the Striped Hamster in spring, autumn and the yearly populations.
Discussion
The relationship between the population and genetic diversity has been studied in some short-term or low-density phases. Evidence from long-term and high density population is short. In this study, 298 samplings were collected and analysed from 2004-2008 in North China Plain. We found that the genetic diversity of the Striped Hamster is density and seasonal dependent. Previous studies showed that fluctuated populations had higher genetic diversity even at low population density [14,17,18].
Elton hypothesis thought considerable reduction in population size could reduce the population genetic diversity. Some studies showed that the genetic diversity changed with the population abundance and they had the same tendency [20], while others researches showed that there also appear opposite tendency between the population abundance and the genetic diversity [33]. In this study, the Striped Hamster population has lower genetic diversity at lowest trap success year (in 2004), which is consistency with the previous studies and the Elton hypothesis.
Fluctuate populations may display temporal oscillation in the amount of genetic diversity [26], while some fluctuate population showed a stable level of genetic diversity during the oscillation period [18,25]. The genetic diversity of the Striped Hamster populations fluctuated with the population abundance during 2004-2008 from Wu Village, Qufu city, Shandong province, China.
Even at the low level of the population size for the cycle vole populations, high effective population sizes and heterozygosity also exist, dispersal may be the important factor to maintain high population genetic variation [19].
Another important hypothesis in studying the population and diversity is Chitty hypothesis which assumes that individuals with aggressive and docile personalities lead to different reproductive strategies in a population [34]. Reproductive strategies directly affect population abundance, and the character of the animal could act on the population fluctuate model. At high population abundance, selection favors aggressive individuals that input less energy into reproduction; at increase period, selection is conducive to the docile individuals which had high reproductive ability. Aggressive and docile characteristics are thought to be genetically inherited. Therefore, we think there is a feedback function that genetic structure of the population could affect the population abundance.
Seasonal change is an important factor affecting population abundance because it determines the living environment such as living temperature and food availability. For example, in summer, rainstorms could destroy the lives burrow of the Striped Hamster and lead to a higher mortality rate. The Striped Hamsters have less and lower quality food, they need more energy to undergoing metabolism and withstanding the low temperatures, which will reduce reproductive activities for the Striped Hamster in winter. While in spring and autumn, the Striped Hamsters have suitable living temperature and adequate food supply, more energy is used for reproductive activities which lead to a high birth rate. Therefore, season is the major factor affecting population abundance for the Striped Hamsters. Similarly, we found the Striped Hamster populations have high population abundance in spring and autumn than summer and winter and the population trap success and genetic diversity were seasonal dependent.
Besides, dispersal pattern [18,35], inbreeding avoidance and genetic drift can affect the genetic diversity of a population and influence its genetic structure [36]. Immigration can slow down the loss of genetic diversity [20], which lead to the accumulation of new alleles and contribute to the increasing of heterozygosity population.
Conclusion
In the Striped Hamster populations, the change tendency in the genetic diversity is in accordance with the population abundance. Genetic diversity of the Striped Hamster is dependent on the population density and the season.
Acknowledgements
This study is support by the National Basic Research Program of China (2007BC109104); National Natural Science Foundation of China (31070332, 31270417, 31300304, 31670385, 31570377).
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- Zhang ZB, Wang ZW. Ecology and management of rodent pests in agriculture. Ocean Beijing 1998.
- Song ZG, Wang DH. Metabolism and thermoregulation in the striped hamster Cricetulus barabensis. J Therm Biol 2003; 28: 509-514.
- Zhao ZJ, Zhu QX, Chen KX, Wang YK, Cao J. Energy budget, behavior and leptin in striped hamsters subjected to food restriction and refeeding. PLoS One 2013; 8: 54244.
- Gyllstr OMM, Hansson L. Dormancy in freshwater zooplankton: induction, termination and the importance of benthic-pelagic coupling. Aquat Sci 2004; 66: 274-295.
- Smith DW, Cooper SD. Competition among cladocera. Ecology 1982; 1982: 1004-1015.
- Rocha O, Matsumura-Tundisi T, Tundisi JEG, Fonseca CP. Predation on and by pelagic Turbellaria in some lakes in Brazil. Intrazooplankton Predation Springer 1990; 1990: 91-101.
- Xu L, Song M, Zhang F, Xu Y, Gao Q. Isolation and characterization of microsatellite markers in the striped hamster (Cricetulus barabensis). Mol Ecol Resour 2008; 8: 1500-1502.
- Ims RA, Andreassen HP. Density-dependent dispersal and spatial population dynamics. Proc Royal Soc B Biol Sci 2005; 272: 913-918.
- Gaines MS, McClenaghan LR. Dispersal in small mammals. Annu Rev Ecol Syst 1980; 11: 163-196.
- Xu L, Xue H, Song M, Zhao Q, Dong J, Liu J, Guo Y, Xu T, Cao X, Wang F, Wang S, Hao S, Yang H, Zhang Z. Variation of genetic diversity in a rapidly expanding population of the greater long-tailed hamster (Tscherskia triton) as revealed by microsatellites. Plos One 2013; 8: 54171.
- Townsend SE, Newey S, Thirgood SJ, Matthews L, Haydon DT. Can parasites drive population cycles in mountain hares? Proc Royal Soc B Biol Sci 2009; 276: 1611-1617.
- Wright S. Evolution and the genetics of populations. Variability within and among natural populations. Univ Chicago Press Chicago IL USA 1978; 4.
- Dong J, Li C, Zhang Z. Density-dependent genetic variation in dynamic populations of the greater long-tailed hamster (Tscherskia triton). J Mammal 2010; 91: 200-207.
- Nor ENK, Angerbj ORA. Genetic perspectives on northern population cycles: bridging the gap between theory and empirical studies. Biol Rev 2014; 89: 493-510.
- Stapley J, Reger J, Feulner PGD, Smadja C, Galindo J, Ekblom R, Bennison C, Ball AD, Beckerman AP, Slate J. Adaptation genomics: the next generation. Trends Ecol Evol 2011; 25: 705-712.
- Karin N, Anders A. Genetic perspectives on northern population cycles: bridging the gap between theory and empirical studies. Biol Rev 2014; 89: 493-510.
- Xie J, Zhang Z. Genetic diversity decreases as population density declines: implications of temporal variation in mitochondrial haplotype frequencies in a natural population of Tscherskia triton. Integr Zool 2006; 1: 188-193.
- Ehrich D, Yoccoz NG, Ims RA. Multi-annual density fluctuations and habitat size enhance genetic variability in two northern voles. Oikos 2009; 118: 1441-1452.
- Rikalainen K, Aspi J, Galarza JA, Koskela E, Mappes T. Maintenance of genetic diversity in cyclic populations-a longitudinal analysis in Myodes glareolus. Ecol Evol 2012; 2: 1491-1502.
- Berthier K, Charbonnel N, Galan M, Chaval Y, Cosson JF. Migration and recovery of the genetic diversity during the increasing density phase in cyclic vole populations. Mol Ecol 2006; 15: 2665-2676.
- Mahy G, Vekemans X, Jacquemart AL. Patterns of allozymic variation within calluna vulgaris populations at seed bank and adult stages. Heredity (Edinb) 1999; 4: 432-440.
- Pemberton JM, Coltman DW, Bancroft DR, Smith JA, Paterson S, Clutton-Brock TH. Molecular genetic variation and selection on genotype. Soay Sheep Dynamics SelectionIsland Popul 2004; 4: 217-242.
- Smith JE, Batzli OG. Dispersal and mortality of prairie voles (Microtus ochrogaster) in fragmented landscapes: a field experiment. Oikos 2006; 112: 209-217.
- Pilot M, Dabrowski MJ, Jancewicz E, Schtickzelle N, Gliwicz J. Temporally stable genetic variability and dynamic kinship structure in a fluctuating population of the root vole Microtus oeconomus. Mol Ecol 2010; 19: 2800-2812.
- Vuorinen JA, Eskelinen O. Long-term stability of allozyme frequencies in a wood lemming, Myopus schisticolor, population with a biased sex ratio and density fluctuations. Heredity (Edinb) 2005; 94: 443-447.
- Motro U, Thomson G. On heterozygosity and the effective size of populations subject to size changes. Evolution 1982; 36: 1059-1066.
- Zhao L, Zhong M, Xue H, Ding J, Wang S, Xu JH, Chen L, Xu LX. Effect of RFRP-3 on reproduction is sex-and developmental status-dependent in the striped hamster (Cricetulus barabensis). Gene 2014; 547: 273-279.
- Berthier K, Galan M, Foltete JC, Charbonnel N, Cosson JF. Genetic structure of the cyclic fossorial water vole (Arvicola terrestris): landscape and demographic influences. Mol Ecol 2005; 14: 2861-2871.
- Kimura M, Crow JF. The Number of alleles that can be maintained in a finite population. Genetics 1964; 49: 725-738.
- Rold A, N-Ruiz I, Dendauw J, Van Bockstaele E, Depicker A, De Loose M. AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol Breeding 2000; 6: 125-134.
- Levene H. On a matching problem in genetics. Sinauer Associates Sunderland Massachusetts Levene2Matching Problem Genetics 1949.
- Cunha-Machado AS, Scarpassa VM. New microsatellite markers for the neotropical malaria vector Anopheles Nuneztovari Sensu Lato 2014; 13: 8856.
- Gaines MS, McClenaghan LR. Rose RK. Temporal patterns of allozymic variation in fluctuating populations of Microtus ochrogaster. Evolution 1978; 32: 723-739.
- Chitty D. The natural selection of self-regulatory behavior in animal populations. ProcEcol Soc Aus 1967; 1967: 1-78.
- Ehrich D, Jorde PE. High genetic variability despite high-amplitude population cycles in lemmings. J Mammal 2005; 86: 380-385.
- Keller LF, Jeffery KJ, Arcese P, Beaumont MA, Hochachka WM, Smith JN, Bruford MW. Immigration and the ephemerality of a natural population bottleneck: evidence from molecular markers. Proc Royal Soc London B Biol Sci 2001; 268: 1387-1394.