Research Article - Biomedical Research (2017) Volume 28, Issue 7
Frequency of single and combined genotypes of GSTM1, GSTT1 and GSTP1 in Mexican individuals: a pilot study
Fernando Mejia-Sanchez1, Maria G. Enríquez-Mejía2, Miriam V. Flores-Merino1, Antonio Laguna Camacho1 and Julieta Castillo-Cadena1*1Laboratorio de Biología Molecular y Cellular, Centro de Investigacion en Ciencias Medicas, Universidad Autonoma del Estado de México, Jesus Carranza No. 205, Col. Universidad, Toluca de Lerdo, México
2Facultad de Química, Universidad Autonoma del Estado de Mexico, Paseo Colon s/n, Col. Universidad, Toluca de Lerdo, Mexico
- *Corresponding Author:
- Julieta Castillo Cadena
Centro de Investigación en Ciencias Médicas
Universidad Autónoma del Estado de México
Toluca de Lerdo, Mexico
Accepted date: November 29, 2016
Abstract
Background: The Glutathione S-transferase is supergene family polymorphic involved in Phase II metabolism. Participate together in the detoxification of xenobiotics. There are few reports where these set of polymorphism of GSTs genes of the same individual are studied. These polymorphisms can be used as biomarkers for predicting disease risk or response to treatment.
Objective: Determine the genotype set of each individual harbouring polymorphisms GSTT1, GSTM1 and GSTP1 in healthy volunteers of Mexican origin.
Methods: We studied 160 samples of healthy volunteers. DNA was extracted from peripheral blood. GSTT1 and GSTM1 polymorphisms were identified by multiplex end point PCR. The identification of GSTP1b (exon 5) and GSTP1c (exon 6) was performed separately by PCR-RFLP.
Results: The frequency for the GSTM1 null was 47%; GSTT1 null 65%; GSTP1 exon 5 for heterozygotes (P1a/P1b) 51.9%, 35.6% in homozygotes (P1b/P1b) and exon 6 heterozygous 25.6% (P1a/ P1c). The most common genotypes set were, wild M1; null T1; a/b P1b; a/a P1c and wild M1; null T1; b/b P1b; a/a P1c, both with a frequency of 12.5%. The null M1; null T1; a/b P1b; a/a P1c frequency was 11.9%. The highest risk genotypes were 3.8% (null M1; null T1; a/b P1b, a/c P1c) and 2.5% (null M1; null T1; b/b P1b; a/c P1c).
Conclusion: The frequencies show genotypes that can be considered high risk, indicating increased susceptibility to xenobiotics. These results are the first report of GSTM1, GSTT1 and GSTP1 combined genotype in Mexican population.
Keywords
Glutathione S-transferase, Polymorphisms, Mexican population
Introduction
About 99.9% of the DNA sequence of the individuals is shared with each other. However, in the remaining 0.1% is where genetic variations are found. These types of variations are known as genetic polymorphisms and they represent different DNA sequences [1,2].
The enzymes of the Glutathione S-Transferase (GST) family consist of several genes that code for a group of isoenzymes involved in Phase II metabolism. The cell can be protected from oxidative damage through the conjugation of glutathione with electrophilic substrates, which generate more soluble compounds [3-5]. The most studied GST subclasses in mammals are Mu (μ), Pi (п) and Theta (θ) [5,6]. The GSTM1 (μ) gene, located on chromosome 1p13.3, consists of eight exons with a length of 4.2 kb and four allelic variants: A (wild), B, C and 0 or null. GSTM1 null is the most frequent polymorphism, which generally result in loss of enzymatic activity [5,7]. On the other hand, the GSTT1 (θ) gene is located on chromosome 22p11.2. It consists of six exons and is flanked by two homologous regions HA3 and HA5. It has two allelic variants: wild GSTT1a and GSTT1-0 or null. This polymorphism is due to a homologous recombination of the HA3 and HA5 regions resulting in a deletion of 5.4 kb [5,8]. The GSTP1 gene (п) is located on chromosome 11q13, it consists of nine exons, with a length of 3.2 kb and it has three allelic variants: GSTP1a, GSTP1b and GSTP1c. The wild allele is GSTP1a and the polymorphic allele GSTP1b is located in exon 5 as a result of the mutation of ATC (Ile) to GTC (Val) at codon 104. The third allele, GSTP1c, also polymorphic, has the same mutation at codon 104. Additionally, it has a second mutation in exon 6, consisting of a base substitution of GCG (Ala) to GTG (Val) [2,9-11].
The GSTs are important because their expression increases significantly in mammalian tumor cells. They have been implicated in resistance to the treatment of different types of cancer drugs [2,3,5,12].
Regularly xenobiotics are metabolized by a group of enzymes that work simultaneously to remove them from the body. The altered enzyme activity by GST polymorphisms is attributed to the GSTT1 and GSTM1 gene deletions, meanwhile for the GSTP1, the SNPs can induce lower enzyme activity [13]. There are several reports in the literature on the frequencies of each of these genes in different populations and also the comparison between them. Nowadays, in literature there are few reports can be found where these set of polymorphism of GSTs genes of the same individual are studied. Most analysis focus on the frequencies of each gene [2,5,11,13-20]. However, the glutathione S-transferase family of enzymes participate together in the detoxification of xenobiotics. Therefore, studying several genes could give a better prognosis of possible susceptibilities in a population. Therefore, the aim of this study was to determine the genotype set of each individual harbouring polymorphisms GSTT1, GSTM1 and GSTP1 in healthy volunteers of Mexican origin.
Material and Methods
Studied group
We recruited participants from January 2014 to January 2015. The participation in this study was voluntary and by invitation. When individuals agreed to take part, they were reported if they had a chronic disease, if they were in treatment or if they were taking any medication. These criteria were used to include apparently healthy individuals. Those who agreed to participate signed a letter of informed consent.
Ethical considerations
The authors certify that all research was done under full compliance with all government policies and the Helsinki Declaration 2013. This study was approved by the Ethics and Research Committee of the Research Center in Medical Sciences at the Autonomous University of the State of Mexico.
DNA extraction
A 4 ml sample of peripheral blood from each participant was collected into a Vacutainer tube with heparin. DNA extraction was performed using a ZR Genomic DNA Kit II (Zymo Research, USA). The products were verified by horizontal electrophoresis in agarose (1%).
Genotyping of GSTT1 and GSTM1
GSTM1 and GSTT1 polymorphisms were identified by multiplex PCR with CYP1A1 gene as control, according to the modified method using specific primers [21]. The mixture had a final volume of 25 μl containing 5l of 5X PCR buffer (Promega), 4 μl of 25 mM MgCl2 (Promega), 1 μl dNTPs, 10 μM (Fermentas), 6.7 μl of molecular biology grade H2O, 0.3 μl of 5 U/μl Taq Polymerase (Promega), 2 μl of DNA template and 1 μl of each primer CYP1A1-f, CYP1A1-r, GSTM1-f, GSTM1-r GSTT1-f, GSTT1-r. All primers had a concentration of 30 pM/μl. The PCR conditions were 5 min at 94°C, 35 cycles of denaturation for 1 min at 94°C, alignment 1 min at 62°C, 1 min extension at 72°C, and a final elongation of 5 min at 72°C. PCR products were verified by horizontal electrophoresis using 1.5% agarose. The determination of the polymorphism of GSTT1 was based on the presence of a 480 bp band, corresponding to wild GSTTl, while its absence denoted a null GSTT1. €Similarly, for GSTM1 the presence of a 215 bp band indicated wild GSTM1 while its absence implied null GSTM1. Finally, a 312 bp band, corresponding to CYP1A1 [21].
Genotyping of GSTP1
Because of GSTP1 polymorphisms are in different regions of the gene, exon 5 (GSTP1b) and exon 6 (GSTP1c), the amplification was performed separately. The PCR-RFLPs was carried out using specific primers for each polymorphism. The mixture had a final volume of 25 μl, containing 5 μl 5X PCR buffer (Promega), 4 μl of 25 mM MgCl2 (Promega), 1 μl dNTPs 10 μM (Fermentas), 10.7 μl of molecular biology grade H2O, 0.3 μl 5 U/μl Taq Polymerase (Promega), 2 μl of DNA template and 1 μl of each primer according to the gene to be amplified GSTP1b-f, GSTP1b-r GSTP1c-f or GSTP1c-r; all primers had a concentration of 30 pM/μl [2]. The same PCR conditions for M1 and T1 were used for GSTP1c genotyping. For GSTP1b, the alignment temperature was 59°C, everything else remained the same.
For exon 5, an amplicon of 176 bp was obtained; while for exon 6 an amplicon of 332 bp. The digestions were performed with enzymes BsmAI (Fermentas) to determine GSTP1b and AciI (Fermentas) to determine GSTP1c. The content of the digestion mixture was: 7 μl H2O (molecular biology grade), 2 μl FastDigest Green buffer (Fermentas), 1 μl Fast-Digest Enzyme (Fermentas) and 10 μl PCR product DNA, for a total volume of 20 μl. Both digestion reactions were incubated at 37°C for 5 minutes. The digestion products were verified by horizontal electrophoresis in 2% of agarose. The identification of polymorphisms was based on the presence of DNA fragments of different sizes. For exon 5, 176 bp, 91 bp and 85 bp fragments correspond to heterozygote P1b, while 91 bp and 85 bp P1b fragments correspond to homozygote and a 176 bp fragment corresponds to wild P1a. Regarding exon 6 polymorphisms, the 332 bp fragment corresponds to homozygote P1c; three fragments, 332 bp, 174 bp and 158 bp correspond to heterozygote P1c and two fragments, 158 bp and 174 bp, to wild P1a [2].
Results
In this study, 160 healthy volunteers participated, 58 were women and 102 were men, representing respectively 36.3% and 63.8% of all cases. The average age was 39.57 years, in a range of 16-72 years.
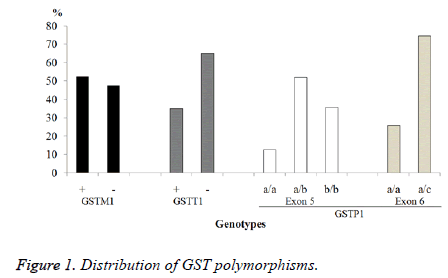
For GSTM1, the number of individuals with gene deletion or null polymorphism was 76 (47.5%), while the wild-type gene was presented in 84 cases representing 52.5%. In the GSTT1 gene, it was found that 104 cases have deletion and 56 had the wild type, 65% and 35% respectively. For GSTP1b gene, the exon 5 and 6 were analysed. In exon 5, it was found: 83 heterozygous (P1a/P1b) 51.9%, 57 homozygous (P1b/P1b) 35.6% and the wild-type gene (P1a/P1a) was identified in 20 individuals (12.5%). For exon 6, it was found that 41 cases had the mutation corresponding to 25.6% of heterozygous (P1a/ P1c) and 119 individuals (74.4%) have the wild-type gene (P1a/P1a), no homozygous was found (Figure 1).
To determine the genotype at each individual, the results of GSTT1, GSTM1 and GSTP1 were grouped. Later, they were identified and grouped individuals according to genotype.
We found that the most common genotype set was: wild GSTM1; null GSTT1; heterozygote (P1a/P1b) exon 5 and wild GSTP1c (P1a/P1a) exon 6 and wild GSTM1; null GSTT1; homozygote (P1b/P1b) exon 5 and wild GSTP1c (P1a/P1a) exon 6, and wild GSTM1; null GSTT1; homozygote (P1a/P1b) exon 5 and wild GSTP1c (P1a/P1a) exon 6, both with a frequency of 12.5%. The genotype with all the genes and the wild alleles: wild GSTM1; wild GSTT1; wild (P1a/P1a) exon 5 and wild GSTP1c (P1a/P1a) exon 6 was present only in the 2.5%. Also, the genotype that confers higher susceptibility to xenobiotics: null GSTM1; null GSTT1; homozygote (P1b/P1b) exon 5 and heterozygote (P1a/P1c) exon 6 had a frequency of 2.5%. The details of these results are presented in Table 1.
| Genotypes | ||||
|---|---|---|---|---|
| GSTM1 | GSTT1 | GSTP1 | GSTP1 | % |
| exon 5 | exon 6 | |||
| + | - | a/b | a/a | 12.5 |
| + | - | b/b | a/a | 12.5 |
| - | - | a/b | a/a | 11.9 |
| - | - | b/b | a/a | 7.5 |
| + | - | a/b | a/c | 7.5 |
| + | + | a/b | a/a | 5.6 |
| + | + | b/b | a/a | 5.6 |
| - | + | a/b | a/a | 5 |
| - | - | a/a | a/a | 4.4 |
| - | - | a/b | a/c | 3.8 |
| - | + | b/b | a/a | 3.1 |
| + | + | a/b | a/c | 3.1 |
| - | - | b/b | a/c | 2.5 |
| - | + | a/a | a/a | 2.5 |
| - | + | a/b | a/c | 2.5 |
| - | + | b/b | a/c | 2.5 |
| + | + | a/a | a/a | 2.5 |
| + | - | a/a | a/a | 1.3 |
| + | - | a/a | a/c | 1.3 |
| + | - | b/b | a/c | 1.3 |
| - | + | a/a | a/c | 0.6 |
| + | + | b/b | a/c | 0.6 |
Table 1. Frequency distribution of genotypes sets.
Discussion
It was found that the frequency for the GSTM1 null polymorphism (47%) in this study is similar to that reported by Montero et al. in a Mexican population, with 42.6 % for the null polymorphism [22]. These results are also consistent with those reported in other populations of the planet. For example, Ye et al. reported a frequency of GSTM1 null genotype of 50% in Caucasian and 51% in Asian and Salah et al. in Tunisian population with a 53.9% for this polymorphism [11,23].
However, the frequencies in this study were higher than the observed in other studies. For instance, Molina et al. and Perez et al. found a frequency of 30% and 37% in Mexican population [24,25]. Investigation in Chile by Quinones et al. and in Africa by Dandara et al. found similar frequencies of 24% and 33 % for GSTM null [19,26]. The present study identified a frequency of 65% for the GSTT1 null polymorphism. Such result differs from findings in other studies. For instance, Perez et al. and Molina et al. reported in Mexican population a frequency of 12%, 15% and 19% [20,24,25]. In Caucasian population, Nelson et al. observed a frequency of 15% and in African population, Dandara et al. and Salah et al. observed a frequency of 25% and 28% [18,19,23].
With respect to the polymorphisms of GSTP1 exon 5, the percentage was 51.9 for heterozygotes (P1a/P1b) and 35.6 in homozygotes (P1b/P1b). These frequencies are similar to the results in Mexican population by Molina et al., Perez et al. and Mejia et al. that report a frequencies for the heterozygote polymorphism P1a/P1b of 50%, 51% and 50% [2,24,25]. In another study in Tunez, Salah et al. the frequency of the heterozygote and homozygote polymorphisms was 54% and 21% respectively [23]. Dandara et al. reported for heterozygote and homozygote polymorphisms of GSTP1 frequencies of 25% and 7% [19]. Similar findings were reported by Ye et al. found a frequency of 10% for Europe, 14% in African Americans and 20% in Asia. However, it is not specified if the polymorphism is heterozygous (P1a/P1b) or homozygous (P1b/P1b) [11]. For polymorphism GSTP1c exon 6, in this work only the heterozygous (P1a/P1c) was identified with a frequency of 25.6%. In contrast, Ye et al. reported only 1% of prevalence. However, they do not specify whether the polymorphism is heterozygous or homozygous [11]. On the other hand, Mejia et al. did not identify any polymorphism in this gene region and reported 100% of the wild allele [2].
With regard to the genotype set of GST, in this study the more frequently genotypes detected were wild GSTM1; null GSTT1; GSTP1 heterozygote exon 5 (P1a/P1b) and GSTP1 wild exon 6 (P1a/P1a). However, in the literature there exist a few studies reporting the genotype set for the GST genes family. Salah et al. found in Tunez population wild GSTM1, wild GSTT1, wild (P1a/P1a) as the most frequent combined genotype [23].
The combined frequencies of genotypes are considered high risk when contain null polymorphisms and SNPs at a greater extent. In contrast, the risk is considered lower when the combined genotype has larger proportion of wild alleles. The present study identified the genotype of higher risk null GSTM1; null GSTT1; GSTP1 homozygote exon 5 (P1b/P1b) and GSTP1 heterozygote exon 6 (P1a/P1c) in 2.5%, and the genotype of lower risk wild GSTM1; wild GSTT1; GSTP1 wild exon 5 (P1a/P1a) and GSTP1wild exon 6 (P1a/P1a) in 2.5%.
Knowing the combined genotype is important because the xenobiotics are metabolized by a set of isoenzymes coded by GST genes. The identification of a single gen and its polymorphisms is not enough to estimate the risk.
The variability of the frequencies can relate to the ancestry of the population and the miscegenation in the different regions of the world. The interbreeding of people of different races differs in Mexico. Caucasian genes predominate in the north of Mexico and genes from mixtures between Spanish people and Mesoamerican natives predominate in the south of Mexico. It is important to consider that there is a big genetic diversity worldwide, which may explain these differences [27].
The results suggested that the frequencies of these polymorphisms could be a handy tool to monitor susceptible groups or populations. Also, the study of the association of polymorphisms can be a useful predictive value of treatment response. In conclusion, these results represent the first combined genotype frequencies reported of GSTM1, GSTT1 and GSTP1 in Mexicans.
Acknowledgements
The authors gratefully acknowledge all participants that volunteered. This study was partially supported by the Autonomous University of the State of Mexico under the agreement 3452/2013 CHT.
References
- Checa CM. Polimorfismos geneticos: Importancia y aplicaciones. Revista del Instituto Nacional de Enfermedades Respiratorias Mexico 2007; 20: 213-221.
- Mejia F, Castillo J, Sanchez J. Development and application in Mexican of a method for the identification of polymorphisms of GSTP1. J Med Medical Sci 2013; 4: 287-290.
- Hayes JD, Pulford DJ. The Glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance Part II. Crit Rev Biochem Mol Biol 1995; 30: 521-600.
- Polimant R, Piacentini S, Hap MF. Map-based study of human soluble Glutathione S-transferase enzymes: The role of natural selection in shaping the single nucleotide polymorphism diversity of xenobiotic-metabolizing genes. Pharmacogenet Genomics 2011; 21: 665-672.
- Rodriguez M, Mejia F, Lecourtois M, Dominguez V, Castillo J. Influence of GSTT1, GSTM1 and GSTP1 polymorphisms on the development of breast cancer. J Cancer Ther 2014; 5: 552-559.
- Strange RC, Spiteri MA, Ramachandran S, Fryer AA. Glutathione-S-transferase family of enzymes. Mutat Res 2001; 482: 21-26.
- Pearson WR, Vorachek WR, Xu SJ. Identification of class-mu glutathione transferase genes GSTM1-GSTM5 on human chromosome 1p13. Am J Human Gene 1993; 53: 220-233.
- Webb G, Vaska V, Coggan M, Board P. Chromosomal localization of the gene for the human theta class glutathione transferase (GSTT1). Genomics 1996; 33: 121-123.
- Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol 2005; 45: 51-88.
- Ali-Osman F, Akande O, Antoun G, Mao JX, Buolamwini J. Characterization and expression in Escherichia coli of full-length cDNAs of three human Glutathione S-Transferase Pi gene variants evidence for differential catalytic activity of the encoded proteins. J Biol Chem 1997; 272: 10004-10012.
- Ye Z, Song H, Higgins JP, Pharoah P, Danesh J. Five glutathione s-transferase gene variants in 23,452 cases of lung cancer and 30,397 controls: meta-analysis of 130 studies. PLoS Med 2006; 3: e91.
- Eaton DL, Bammler TK. Concise review of the glutathione S-transferases and their significance to toxicology. Toxicol Sci 1999; 49: 156-164.
- Soto QO, Cabrera GP, Tellez TG, Barrera FJL, Juarez RA, Castillo CJ. Relationship of polymorphisms of Glutathione S-Transferase GSTT1 and GSTM1 with the response to chemotherapy in Mexican women with advanced breast cancer. J Cancer Ther 2011; 2: 354-361.
- Salimi S, Nakhaee A, Jafari M, Jahantigh D, Sandooghi M. Combination effect of GSTM1, GSTT1 and GSTP1 polymorphisms and risk of systemic lupus erythematosus. Iran J Public Health 2015; 44: 814-821.
- Jia W, Sun JY, Jia KY, Liu XC. Role of GSTM1, GSTT1, and GSTP1 IIe105Val gene polymorphisms in the response to chemotherapy and overall survival of advanced non-small cell lung cancer. Genet Mol Res 2016; 15.
- Kumara A, Yadava A, Kumar GS, Dev K, Gulati S, Kumar GS, Gupta R, Aggarwal N. Allelic variation of GSTM1 and GSTT1 genes in Haryana population. Genomic Med Biomark Health Sci 2012; 4: 98-102
- Pemble S, Schroeder KR, Spencer SR, Meyer DJ, Hallier E, Bolt HM, Ketterer B, Taylor JB. Human Glutation S-transferasa theta (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem J 1994; 300: 271-276.
- Nelson HH, Wiencke JK, Christiani DC, Cheng TJ, Zuo ZF, Schwartz BS, Lee BK, Spitz MR, Wang M, Xu X. Ethnic differences in the prevalence of the homozygous deleted genotype of glutathione S-transferase theta. Carcinogen 1995; 16: 1243-1246.
- Dandara C, Sayi J, Masimirembwa CM, Magimba A, Kaaya S, De Sommers K, Snyman JR, Hasler JA. Genetic polymorphism of cytochrome P450 1A1 (Cyp1A1) and glutathione transferases (M1, T1 and P1) among Africans. Clin Chem Lab Med 2002; 40: 952-957.
- Perez-Morales R, Castro-Hernandez C, Gonsebatt ME, Rubio J. Polymorphism of CYP1A1*2C, GSTM1*0, and GSTT1*0 in a Mexican Mestizo population: a similitude analysis. Hum Biol 2008; 80: 457-465.
- Abdel-Rahman SZ, el-Zein RA, Anwar WA, Au WW. A multiplex PCR procedure for polymorphic analysis of GSTM1 and GSTT1 genes in population studies. Cancer Lett 1996; 107: 229-233.
- Montero R, Araujo A, Carranza P, Mejía-Loza V, Serrano L. Genotype frequencies of polymorphic GSTM1, GSTT1, and cytochrome P450 CYP1A1 in Mexicans. Hum Biol 2007; 79: 299-312.
- Salah GB, Kallabi F, Maatoug S, Mkaouar-Rebai E, Fourati A, Fakhfakh F, Ayadi H, Kamoun H. Polymorphisms of Glutation S-transferases M1, T1, P1, and A1 genes in the Tunisian population: An intra and interethnic comparative approach. Gene 2012; 498: 317-322.
- Molina E, Perez-Morales R, Rubio J, Petrosyan P, Cadena LH. The GSTM1null (deletion) and MGMT84 rs12917 (Phe/Phe) haplotype are associated with bulky DNA adduct levels in human leukocytes. Mutat Res 2013; 758: 62-68.
- Perez-Morales R, Mendez-Ramirez I, Moreno-Macias H, Mendoza-Posadas AD, Martinez-Ramirez OC. Genetic susceptibility to lung cancer based on candidate genes in a sample from the Mexican Mestizo population: a case-control study. Lung 2014; 192: 167-173.
- Quinones L, Lucas D, Godoy J, Caceres D, Berthou F, Varela N, Lee K, Acevedo C, Martinez L, Aguilera AM, Gil L. CYP1A1, CYP2E1 and GSTM1 genetic polymorphisms. The effect of single and combined genotypes on lung cancer susceptibility in Chilean people. Cancer Lett 2001; 174: 35-44.
- Moreno EA, Gignoux CR, Fernandez LJC, Zakharia F, Sikora M, Contrera AV, Acuna AV, Sandoval K, Eng C, Romero HS, Ortiz TP, Robles V, Kenny EE, Nuno AI, Barquera LR, Macin PG, Granados AG, Huntsman S, Galanter SJ, Via M, Ford JG, Chapela R, Rodriguez CW, Rodriguez SJR, Romieu I, Sienra MJJ, Navarro B, London SJ, Ruiz LA, Garcia HR, Estrada K, Hidalgo MA, Jimenez SG, Carnevale A, Soberon X, Canizales QS, Rangel VH, Silva ZI, Gonzalez BE, Bustamante CD. Human genetics. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science 2014; 344: 1280-1285.