Research Article - Biomedical Research (2017) Volume 28, Issue 22
Expression and functions of microRNA-139 in osteosarcoma
Qi Liao*, Jianghua Ming, Geliang Hu, Yaming Li, Chun Zhang and Yanchao Gu
Department of Orthopedics, Renmin Hospital of Wuhan University, Wuhan, PR China
- *Corresponding Author:
- Qi Liao
Department of Orthopedics
Renmin Hospital of Wuhan University, PR China
Accepted date: October 14, 2017
Abstract
Osteosarcoma (OS), derived from the primitive bone-forming mesenchymal cells in the long bones, is the leading cause of cancer-related deaths in adolescents and children. MiRcroRNA-139 (miR-139) has been reported to be abnormally expressed and play important roles in several cancer types. However, the expression pattern, detailed biological roles and associated molecular mechanisms of miR-139 in OS have remained to be fully elucidated. This study showed that miR-139 expression was downregulated in OS tissues and cell lines. Upregulation of miR-139 significantly inhibited cell proliferation and invasion in OS. Bioinformatics analysis predicted that ROCK1 is a potential target of miR-139. Luciferase reporter assay demonstrated that miR-139 can directly interact with the 3’-untranslated region of ROCK1. RT-qPCR and Western blotting analysis demonstrated that miR-139 had regulatory effects on endogenous ROCK1 expression in OS. Furthermore, ROCK1 knockdown significantly produced effects similar to those induced by miR-139 overexpression in OS cells. Therefore, the miR-139/ROCK1 axis may be a potential target for novel potential therapeutic strategies for treatment of OS.
Keywords
microRNA-139, Proliferation, Invasion, ROCK1, Osteosarcoma
Introduction
Osteosarcoma (OS), derived from primitive bone-forming mesenchymal cells in long bones, is the leading cause of cancer-related deaths in adolescents and children [1]. Current major treatments for patients with OS include surgical resection, chemotherapy and radiotherapy [2]. Despite great efforts regarding treatments of OS, the prognosis of this disease remains poor mainly because of recurrence and metastasis [3]. The 5 y overall survival rates are 60%-70% and less than 30% for patients with OS at locally advanced stage and patients who present with metastatic disease, respectively [4]. Therefore, the molecular mechanisms underlying OS formation and progression must be elucidated to develop novel treatment regimens for patients with this disease.
MicroRNAs (miRNAs) are a large family of noncoding, endogenous and short RNA molecules with approximately 19-25 nucleotides in length [5]. MiRNAs negatively regulate gene expression by directly binding to the 3’-Untranslated Regions (UTRs) of their corresponding genes, leading to cleavage and repressed translation of their target mRNAs [6]. Thus far, more than 1,000 miRNAs have been identified in the human genome; these miRNAs may regulate the expression of approximately 30% of all protein-coding genes [7]. A number of evidence indicated that miRNAs are frequently aberrantly expressed in various types of cancer, including OS [8]. Deregulated miRNAs participate in the regulation of tumourigenesis and tumour development through regulating a large number of biological processes, including cell proliferation, cell cycle, apoptosis, invasion, angiogenesis and metastasis [9]. Hence, miRNAs may serve as potential therapeutic targets for anticancer therapy.
MiR-139 has been reported to be abnormally expressed and play important roles in several types of cancer. However, the expression pattern, detailed biological roles and associated molecular mechanisms of miR-139 in OS remain to be fully elucidated. Therefore, this study aims to detect the expression level of miR-139 in OS and investigate its effects on OS as well as its underlying molecular mechanisms.
Material and Methods
Clinical specimens
This research was approved by the Ethics Committee of Renmin Hospital of Wuhan University. In addition, written informed consent was obtained from all participator. A total of 23 primary OS tissues and corresponding adjacent normal tissues were collected at Renmin Hospital of Wuhan University. None of these OS patients had been treated with radiation therapy or chemotherapy before surgery. All tissue specimens were snap-frozen in liquid nitrogen and stored at -80°C until further use.
Cell lines and transfection
The human normal osteoblasts (hFOB1.19), three OS cell lines (U2OS, Saos-2, and MG-63) and HEK293T cell line were acquired from Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). All cell lines were cultured in RPMI-1640 medium containing 10% fetal bovine serum(FBS), 100 U/ml penicillin and 100 U/ml streptomycin (all Gibco, Grand Island, NY, USA), at 37°C in a humidified atmosphere of 5% CO2.
MiR-139 mimics, negative control miRNA mimics (miR-NC), a small interfering RNA (siRNA) targeting ROCK1 (ROCK1 siRNA) and negative control siRNA (NC siRNA) were chemically synthesized by RiboBio (Guangzhou, China). Cell transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissues or cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Waltham, MA, USA). Reverse transcription was performed to synthesize cDNA using a PrimeScript RT Reagent kit (Takara Bio, Inc., Otsu, Japan). To quantify miR-139 expression, quantitative PCR was conducted with a SYBR Premix Ex Taq™ kit (Takara Biotechnology Co., Ltd., Dalian, China). SYBR Green PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Waltham, MA, USA) was adopted to detect ROCK1 mRNA expression. U6 snRNA and GAPDH were used as internal control for miR-139 and ROCK1 mRNA, respectively. Relative expression was calculated using the 2-ΔΔCq method.
MTT assay
Transfected cells were collected 24 h post-transfection and seeded into 96-well plates at a density of 3 × 103 cells in each well. After incubation at 37°C for 0, 24, 48 or 72 h, 20 μl of MTT reagent (5 mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added into each well and incubated at 37°C for an additional 4 h. Afterwards, 150 μl of dimethyl sulfoxide was added to each well and incubated at 37°C for another 10 min. Finally, absorbance at 490 nm was examined using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Each assay was performed in triplicate and repeated three times.
Cell invasion assay
Cell invasion assay was conducted to evaluate cell invasion ability by using Transwell chambers coated with Matrigel (BD Biosciences, San Jose, CA, USA). A total of 1 × 105 transfected cells were suspended in 200 μl of FBS-free RPMI-1640 medium and seeded in the upper chamber. The lower chamber was filled with 500 μl of RPMI-1640 medium containing 20% FBS. After incubation at 37°C for 24 h, noninvading cells were removed carefully by using cotton swabs. The invaded cells were fixed with 100% methanol, stained with 0.5% crystal violet, washed with PBS and dried in air. Five fields were randomly selected, and the number of invaded cells was counted under an inverted microscope (X200; Olympus, Tokyo, Japan). The experiments were performed in triplicate.
Luciferase reporter assay
Luciferase plasmid, pMIR-ROCK1 -3'-UTR Wild-type (Wt) and pMIR-ROCK1-3'-UTR Mutant (Mut), were chemically synthesized and confirmed by GenePharma Co., Ltd (Shanghai, China). HEK293T cells were seeded into 24-well plates at a density of 50%-60% confluence. After incubation overnight, miR-139 mimics or miR-NC was transfected into HEK293T cells together with pMIR-ROCK1-3'-UTR Wt or pMIR-ROCK1-3'-UTR Mut using Lipofectamine 2000. At 48 h post-transfection, a Dual-Luciferase Reporter Assay system (Promega, Madison, WI, USA) was used to detect the luciferase activities.
Western blotting analysis
Total protein was extracted from tissues or cells with ice-cold RIPA buffer (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Equal quantity of protein was separated by 10% SDS-PAGE, transferred to a polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA, USA), and blocked with 5% non-fat dry milk in TBST at room temperature for 2 h. Subsequently, the membranes were incubated overnight at 4°C with primary antibodies against ROCK1 (sc-374388) or GAPDH (sc-32233; both from Santa Cruz Biotechnology, CA, USA). After being washed three times with TBST, the membranes were probed with corresponding horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology, CA, USA) at room temperature for 1 h. The protein bands were then visualized using the ECL Western Blotting kit (Pierce Biotechnology, Inc., Rockford, IL, USA).
Statistical analysis
Data are shown as the mean ± standard deviation. The statistical difference between two groups was assessed by using student’s t-test or one-way ANOVA. P value less than 0.05 was considered statistically significant.
Results
MiR-139 expression is downregulated in OS tissues and cell lines
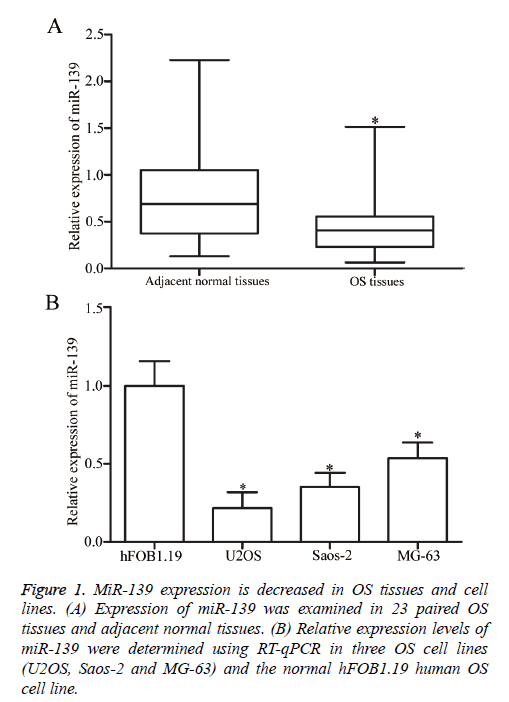
RT-qPCR analysis was performed to determine whether miR-139 is differentially expressed in OS. MiR-139 expression was detected in 23 paired OS tissues and adjacent normal tissues. The results showed that miR-139 was significantly downregulated in OS tissues compared with that in adjacent normal tissues (P<0.05, Figure 1A). Moreover, the expression levels of miR-139 in human normal osteoblasts (hFOB1.19) and three OS cell lines (U2OS, Saos-2 and MG-63) were examined using RT-qPCR analysis. As shown in Figure 1B, miR-139 expression was lower in all three OS cell lines than that in hFOB1.19. Hence, miR-139 may play important roles in OS initiation and progression.
Figure 1: MiR-139 expression is decreased in OS tissues and cell lines. (A) Expression of miR-139 was examined in 23 paired OS tissues and adjacent normal tissues. (B) Relative expression levels of miR-139 were determined using RT-qPCR in three OS cell lines (U2OS, Saos-2 and MG-63) and the normal hFOB1.19 human OS cell line.
MiR-139 inhibits the proliferation and invasion of OS cells
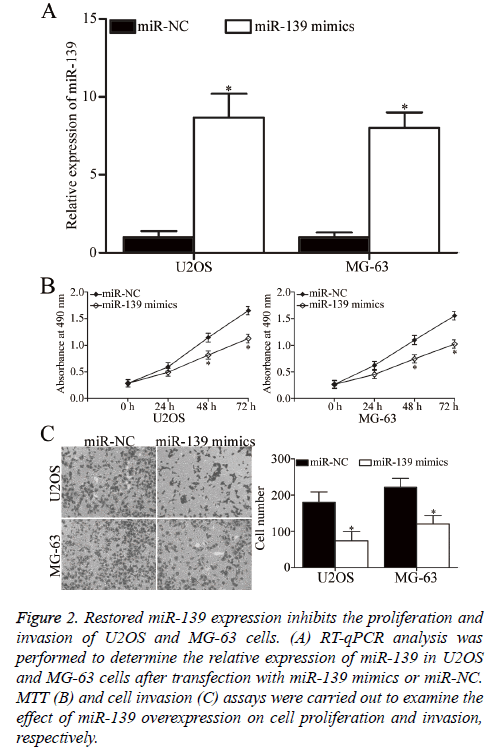
U2OS and MG-63 cells, which expressed relatively low miR-139 expression, were transfected with miR-139 mimics to increase its expression and explore the role of miR-139 in OS. After transfection, RT-qPCR analysis confirmed that miR-139 was markedly upregulated in U2OS and MG-63 cells transfected with miR-139 mimics (P<0.05, Figure 2A). MTT assay was then performed to investigate the effect of miR-139 overexpression on OS cell proliferation. As shown in Figure 2B, restoration of miR-139 expression inhibited the proliferation of U2OS and MG-63 cells (P<0.05, Figure 2B). Additionally, cell invasion assay was utilised to evaluate the invasion ability in U2OS and MG-63 cells transfected with miR-139 mimics or miR-NC. The cell invasion capacity decreased in the group with overexpressed miR-139 compared with that in the miR-NC group (P<0.05, Figure 2C). Hence, miR-139 may act as a tumour suppressor in OS.
Figure 2: Restored miR-139 expression inhibits the proliferation and invasion of U2OS and MG-63 cells. (A) RT-qPCR analysis was performed to determine the relative expression of miR-139 in U2OS and MG-63 cells after transfection with miR-139 mimics or miR-NC. MTT (B) and cell invasion (C) assays were carried out to examine the effect of miR-139 overexpression on cell proliferation and invasion, respectively.
ROCK1 is a direct target of miR-139 in OS
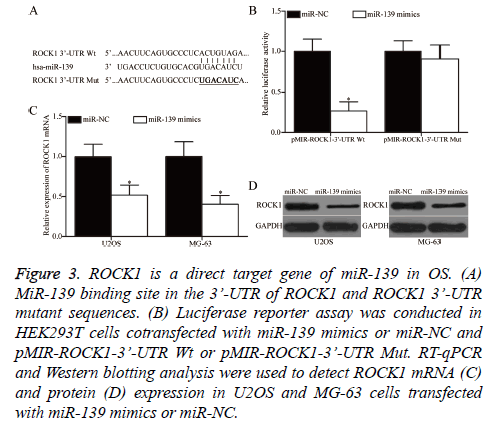
Bioinformatics analysis was performed to predict the potential targets and study the mechanism underlying the tumoursuppressive roles of miR-139 in OS. ROCK1 was predicted as a target of miR-139 and was selected for further confirmation (Figure 3A). Luciferase reporter assay was conducted to determine whether miR-139 could directly interact with the 3’- UTR of miR-139. HEK293T cells were cotransfected with miR-139 mimics or miR-NC and pMIR-ROCK1-3’-UTR Wt or pMIR-ROCK1-3’-UTR Mut. As shown in Figure 3B, miR-139 overexpression significantly decreased the luciferase activity of pMIR-ROCK1-3’-UTR Wt, but not that of pMIRROCK1- 3’-UTR Mut, in HEK293T cells (P<0.05). Finally, RT-qPCR and Western blotting analysis were conducted to investigate whether miR-139 exerts regulatory effects on endogenous ROCK1 expression in OS. As presented in Figures 3C and 3D, the upregulation of miR-139 reduced ROCK1 expression in U2OS and MG-63 cells at the mRNA (P<0.05) and protein (P<0.05) levels. Overall, ZEB1 is a direct target of miR-139 in OS.
Figure 3: ROCK1 is a direct target gene of miR-139 in OS. (A) MiR-139 binding site in the 3’-UTR of ROCK1 and ROCK1 3’-UTR mutant sequences. (B) Luciferase reporter assay was conducted in HEK293T cells cotransfected with miR-139 mimics or miR-NC and pMIR-ROCK1-3’-UTR Wt or pMIR-ROCK1-3’-UTR Mut. RT-qPCR and Western blotting analysis were used to detect ROCK1 mRNA (C) and protein (D) expression in U2OS and MG-63 cells transfected with miR-139 mimics or miR-NC.
Knockdown of ROCK1 produces similar effects to those induced by miR-139 overexpression in OS cells
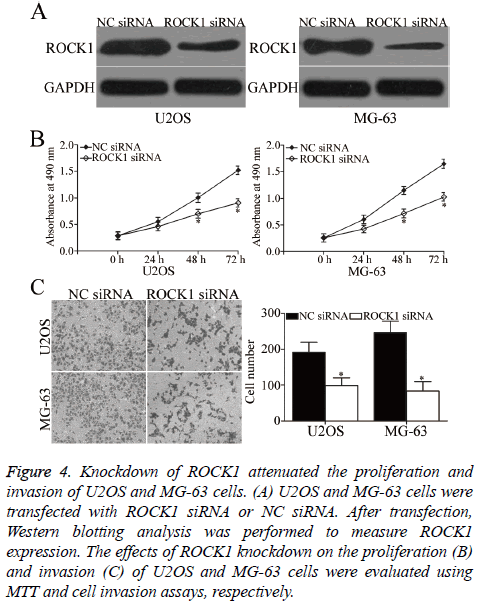
To investigate whether ROCK1 contributes to the suppressive effects of miR-139 in OS, we knocked down ROCK1 expression by siRNA in U2OS and MG-63 cells. After transfection, Western blotting analysis was carried out to evaluate transfection efficiency. ROCK1 was obviously downregulated in ROCK1 siRNA-transfected U2OS and MG-63 cells compared with that in cells transfected with NC siRNA (P<0.05, Figure 4A). Subsequent functional assays demonstrated that similar to miR-139 overexpression, ROCK1 under expression suppressed the proliferation (P<0.05, Figure 4B) and invasion (P<0.05, Figure 4C) of U2OS and MG-63 cells. Hence, miR-139 may play tumour-suppressing roles in OS, at least in part, by regulating ROCK1.
Figure 4: Knockdown of ROCK1 attenuated the proliferation and invasion of U2OS and MG-63 cells. (A) U2OS and MG-63 cells were transfected with ROCK1 siRNA or NC siRNA. After transfection, Western blotting analysis was performed to measure ROCK1 expression. The effects of ROCK1 knockdown on the proliferation (B) and invasion (C) of U2OS and MG-63 cells were evaluated using MTT and cell invasion assays, respectively.
Discussion
Numerous studies have reported that miR-139 is abnormally expressed in various types of human cancers. For example, miR-139 is downregulated in colorectal cancer tissues and cell lines. Low miR-139 expression is associated with tumour stage, progression and metastasis [10,11]. Patients with colorectal cancer and low miR-139 expression showed poorer prognosis than those with high miR-139 expression [12]. In breast cancer, the expression level of miR-139 was lower in tumour tissues than that in normal tissues. Pronounced downregulation of miR-139 expression was detected in breast cancer tissues that showed high grade, estrogen receptor negative, progesterone receptor negative, high proliferation rate or large size [13]. In hepatocellular carcinoma, miR-139 was lowly expressed in tumour tissues. Decreased miR-139 expression is associated with venous invasion, microsatellite formation and tumour encapsulation and differentiation [14]. Downregulation of miR-139 was also observed in laryngeal squamous carcinoma [15], cervical cancer [16], bladder cancer [17,18] and glioma [19]. These findings suggested that miR-139 may serve as a biomarker in these types of human cancer.
MiR-139 is implicated in the initiation and progression of human cancers. For instance, in colorectal cancer, upregulation of miR-139 inhibited cell proliferation, migration, invasion, metastasis and epithelial-mesenchymal transition; promoted apoptosis in vitro; and decreased tumour growth in vivo [10-12,20]. Zhang et al. reported that miR-139 overexpression attenuated cell proliferation, viability and motility; increased apoptosis; and induced cell cycle arrest in the S phase in breast cancer [21,22]. Huang et al. found that ectopic expression of miR-139 promoted apoptosis and repressed metastasis in cervical cancer [16]. Luo et al. revealed that restoration of miR-139 expression suppressed cell proliferation, migration, invasion and self-renewal in bladder cancer [17,18]. Dai et al. reported that miR-139 played tumour suppressive roles in glioma by regulating cell proliferation, apoptosis, invasion, metastasis and chemosensitivity [19,23]. Qiu et al. indicated that restoration of miR-139 expression reduced cell proliferation, metastasis and epithelial-mesenchymal transition in vitro and decreased the incidence and severity of lung metastasis from orthotopic liver tumours in mice [14,24,25]. These findings suggested that miR-139 may provide potential therapeutic targets for treatments of these human cancers.
Bioinformatics analysis was performed to predict the target genes of miR-139, especially ROCK1, and illustrate the potential molecular mechanism through which miR-139 inhibits tumour growth and metastasis in OS. Luciferase reporter assay demonstrated that the 3’-UTR of ROCK1 could be directly targeted by miR-139. RT-qPCR and Western blotting analysis verified that miR-139 exerted a negative regulatory effect on ROCK1 expression at the mRNA and protein levels. ROCK1 knockdown showed similar effects to that of miR-139 expression in cell proliferation and invasion in OS. Hence, ROCK1 is a direct and functional target of miR-139 in OS.
ROCK1, located at chromosome 18 (18q11.1) [26], is upregulated in different types of human cancers, such as breast cancer [27], gastric cancer [28], glioma [29] and lung cancer [30]. The expression level of ROCK1 evidently increased in OS tissues and cell lines. Upregulation of ROCK1 was associated with poor outcomes for patients with OS. Functional experiment demonstrated that ROCK1 knockdown suppressed cell proliferation and viability and enhanced apoptosis in OS. Other studies also reported that ROCK1 was involved in the growth and metastasis of OS [31-33]. Hence, ROCK1 may be an effective therapeutic target for anti-tumour therapy of patients with OS.
In conclusion, this study demonstrated that miR-139 inhibited cell proliferation and invasion in OS, at least, partially by regulating ROCK1. Hence, miR-139 may be a novel effective therapeutic target for OS treatments in the future.
References
- Guillon MA, Mary PM, Brugiere L, Marec-Berard P, Pacquement HD, Schmitt C, Guinebretiere JM, Tabone MD. Clinical characteristics and prognosis of osteosarcoma in young children: a retrospective series of 15 cases. BMC Cancer 2011; 11: 407.
- Chou AJ, Geller DS, Gorlick R. Therapy for osteosarcoma: where do we go from here? Paediatr Drugs 2008; 10: 315-327.
- Thompson LD. Osteosarcoma. Ear Nose Throat J 2013; 92: 288-290.
- PosthumaDeBoer J, Witlox MA, Kaspers GJ, van Royen BJ. Molecular alterations as target for therapy in metastatic osteosarcoma: a review of literature. Clin Exp Metastasis 2011; 28: 493-503.
- Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet 2008; 9: 831-842.
- Ambros V. The functions of animal microRNAs. Nature 2004; 431: 350-355.
- Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet 2007; 8: 93-103.
- Li CH, Yu TB, Qiu HW, Zhao X. miR-150 is downregulated in osteosarcoma and suppresses cell proliferation, migration and invasion by targeting ROCK1. Oncol Lett 2017; 13: 2191-2197.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281-297.
- Shen K, Liang Q, Xu K, Cui D, Jiang L, Yin P, Lu Y, Li Q, Liu J. MiR-139 inhibits invasion and metastasis of colorectal cancer by targeting the type I insulin-like growth factor receptor. Biochem Pharmacol 2012; 84: 320-330.
- Li Q, Liang X, Wang Y, Meng X, Xu Y, Cai S, Wang Z, Liu J, Cai G. miR-139-5p inhibits the epithelial-mesenchymal transition and enhances the chemotherapeutic sensitivity of colorectal cancer cells by downregulating BCL2. Sci Rep 2016; 6: 27157.
- Song M, Yin Y, Zhang J, Zhang B, Bian Z, Quan C, Zhou L, Hu Y, Wang Q, Ni S, Fei B, Wang W, Du X, Hua D, Huang Z. MiR-139-5p inhibits migration and invasion of colorectal cancer by downregulating AMFR and NOTCH1. Protein Cell 2014; 5: 851-861.
- Dai H, Gallagher D, Schmitt S, Pessetto ZY, Fan F. Role of miR-139 as a surrogate marker for tumor aggression in breast cancer. Hum Pathol 2017; 61: 68-77.
- Wong CC, Wong CM, Tung EK, Au SL, Lee JM. The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating Rho-kinase 2. Gastroenterology 2011; 140: 322-331.
- Luo HN, Wang ZH, Sheng Y, Zhang Q, Yan J, Hou J, Zhu K, Cheng Y, Xu YL, Zhang XH, Xu M, Ren XY. MiR-139 targets CXCR4 and inhibits the proliferation and metastasis of laryngeal squamous carcinoma cells. Med Oncol 2014; 31: 789.
- Huang P, Xi J, Liu S. MiR-139-3p induces cell apoptosis and inhibits metastasis of cervical cancer by targeting NOB1. Biomed Pharmacother 2016; 83: 850-856.
- Luo H, Yang R, Li C, Tong Y, Fan L. MicroRNA-139-5p inhibits bladder cancer proliferation and self-renewal by targeting the Bmi1 oncogene. Tumour Biol 2017; 39: 1010428317718414.
- Yonemori M, Seki N, Yoshino H, Matsushita R, Miyamoto K, Nakagawa M, Enokida H. Dual tumor-suppressors miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in bladder cancer. Cancer Sci 2016; 107: 1233-1242.
- Dai S, Wang X, Li X, Cao Y. MicroRNA-139-5p acts as a tumor suppressor by targeting ELTD1 and regulating cell cycle in glioblastoma multiforme. Biochem Biophys Res Commun 2015; 467: 204-210.
- Zhang L, Dong Y, Zhu N, Tsoi H, Zhao Z, Wu CW, Wang K, Zheng S, Ng SS, Chan FK, Sung JJ, Yu J. microRNA-139-5p exerts tumor suppressor function by targeting NOTCH1 in colorectal cancer. Mol Cancer 2014; 13: 124.
- Zhang HD, Sun DW, Mao L, Zhang J, Jiang LH, Li J, Wu Y, Ji H, Chen W, Wang J, Ma R, Cao HX, Wu JZ, Tang JH. MiR-139-5p inhibits the biological function of breast cancer cells by targeting Notch1 and mediates chemosensitivity to docetaxel. Biochem Biophys Res Commun 2015; 465: 702-713.
- Hua W, Sa KD, Zhang X, Jia LT, Zhao J, Yang AG, Zhang R, Fan J, Bian K. MicroRNA-139 suppresses proliferation in luminal type breast cancer cells by targeting Topoisomerase II alpha. Biochem Biophys Res Commun 2015; 463: 1077-1083.
- Li RY, Chen LC, Zhang HY, Du WZ, Feng Y. MiR-139 inhibits Mcl-1 expression and potentiates TMZ-induced apoptosis in glioma. CNS Neurosci Ther 2013; 19: 477-483.
- Gu W, Li X, Wang J. miR-139 regulates the proliferation and invasion of hepatocellular carcinoma through the WNT/TCF-4 pathway. Oncol Rep 2014; 31: 397-404.
- Qiu G, Lin Y, Zhang H, Wu D. miR-139-5p inhibits epithelial-mesenchymal transition, migration and invasion of hepatocellular carcinoma cells by targeting ZEB1 and ZEB2. Biochem Biophys Res Commun 2015; 463: 315-321.
- Lock FE, Ryan KR, Poulter NS, Parsons M, Hotchin NA. Differential regulation of adhesion complex turnover by ROCK1 and ROCK2. PLoS One 2012; 7: 31423.
- Bottino J, Gelaleti GB, Maschio LB, Jardim-Perassi BV, de Campos Zuccari DA. Immunoexpression of ROCK-1 and MMP-9 as prognostic markers in breast cancer. Acta Histochem 2014; 116: 1367-1373.
- Wu YJ, Tang Y, Li ZF, Li Z, Zhao Y. Expression and significance of Rac1, Pak1 and Rock1 in gastric carcinoma. Asia Pac J Clin Oncol 2014; 10: 33-39.
- Wan X, Cheng Q, Peng R, Ma Z, Chen Z. ROCK1, a novel target of miR-145, promotes glioma cell invasion. Mol Med Rep 2014; 9: 1877-1882.
- Vigil D, Kim TY, Plachco A, Garton AJ, Castaldo L, Pachter JA, Dong H, Chen X, Tokar B, Campbell SL, Der CJ. ROCK1 and ROCK2 are required for non-small cell lung cancer anchorage-independent growth and invasion. Cancer Res 2012; 72: 5338-5347.
- Zhang S, Zhao Y, Wang L. MicroRNA-198 inhibited tumorous behaviors of human osteosarcoma through directly targeting ROCK1. Biochem Biophys Res Commun 2016; 472: 557-565.
- Huang J, Shi Y, Li H, Yang M, Liu G. MicroRNA-144 acts as a tumor suppressor by targeting Rho-associated coiled-coil containing protein kinase 1 in osteosarcoma cells. Mol Med Rep 2015; 12: 4554-4559.
- Lei P, Xie J, Wang L, Yang X, Dai Z. microRNA-145 inhibits osteosarcoma cell proliferation and invasion by targeting ROCK1. Mol Med Rep 2014; 10: 155-160.