Research Article - Biomedical Research (2018) Volume 29, Issue 1
Retracted: Exploring key genes and pathways underlying metastasis endometrial cancer based on gene expression microarray
Hui Wang, Xinhui Sun, Shuai Shao, Na Liu*Department of Obstetrics and Gynecology, People’s Hospital of Linyi City, Shandong Province, PR China
- *Corresponding Author:
- Na Liu
Department of Obstetrics and Gynecology
People’s Hospital of Linyi City, PR China
Accepted date: October 24, 2017
DOI: 10.4066/biomedicalresearch.29-17-1326
Visit for more related articles at Biomedical ResearchAbstract
Purpose: To reveal the key genes and pathways involved in the metastasis of endometrial cancer remains.
Methods: Microarray data GSE29436 consisting of 4 progressive endometrial cancer samples and 4 nonprogressive controls were downloaded from Gene Expression Omnibus database. Differentially Expressed Genes (DEGs) were screened out using Limma package in R, followed by hierarchical clustering. Gene Ontology and pathway enrichment analysis were performed for DEGs.
Results: Bioinformatic analysis revealed a total of 65 DEGs between progressive and non-progressive samples. Functional annotation showed that those genes were mainly enriched in functions of cell proliferation, MPKA and TGF-beta signaling pathways, which involved with up-regulated genes pleiomorphic adenoma gene 1 (PLAG1), Insulin-Like Growth Factor 2 (IGF2), and down-regulated genes Fibroblast Growth Factor 20 (FGF20) and Thrombospondin 4 (THBS4).
Conclusion: Sixty-five identified genes in progressive endometrial cancer samples were mainly associated with tumor metastasis possibly by enhancing cell proliferation and affecting MAPK and TGF-beta signaling pathways. High expression of PLAG1, IGF2 and low expression of FGF20, THBS4, therefore, appear to play an important role in tumorigenesis and progression of endometrial cancer.
Keywords
Endometrial cancer, Differentially expressed genes, Pathways, Microarray.
Introduction
Endometrial cancer is the sixth most common cancer affecting women in the western world, with at least 320,000 new cases being diagnosed resulting in 74,000 disease-related deaths in women worldwide [1]. Although endometrial cancer is efficiently diagnosed and successfully treated, the treatment of aggressive and progesterone-resistant cancer is difficult. However, patients with early stage disease have 5 y survival rates over 80% but 15-20% develops metastasis [2].
There are two types of endometrial cancer (types I and II) with different molecular expression profiles and clinical and histopathological behaviors [3,4]. Type I endometrial cancer is estrogen-dependent with endometrioid morphology and accounts for 75% of endometrial cancer [5]. Type II cancer exhibits serious histological alterations and poorly differentiated endometrioid morphology with extra-uterine spread and myometrial invasion. The mechanisms of endometrial cancer metastasis have been partly elucidated, but not fully understood.
In the current study, we aimed to further elucidate the molecular mechanism of endometrial cancer metastasis and provide potential therapeutic target for treating endometrial cancer. Based on the microarray data GSE29436, further analysis was performed by identifying Differentially Expressed Genes (DEGs) in progressive samples compared with nonprogressive control, as well as functional annotation of DEGs. Finally, functions and involved biological pathway associated with metastasis of endometrial cancer were detected.
Materials and Methods
Data source
Microarray data GSE29436 were downloaded from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database based on the platform of Affymetrix Human Genome U133 Plus 2.0 Array. The dataset included 4 progressive endometrial cancer specimen and 4 non-progressive controls.
Data preprocessing and identification of DEGs
The affy package in R [6] was used to process the CEL files by converting the raw probe intensities to probe set expression values. Raw data were background-corrected and normalized with Robust Multi-Array Average (RMA) procedure [7]. Replicate probes for one gene were summarized by mean value as expression value for one gene. To identify critical genes in endometrial cancer metastasis, DEGs were detected using limma package in R [8] by comparing the expression profile between progressive and non-progressive samples. Batch effects in microarray expression data were adjusted using empirical Bayes method [9]. Genes were considered to be differentially expressed in progressive samples if |log2FC (fold change)|>1 and adjusted P-value<0.01. Then, hierarchical clustering was carried out for DEGs based on expression value of each sample.
Functional annotation of DEGs in progressive cancer
Gene Ontology (GO) database [10] includes functions from three categories, biological process, cellular component and molecular function. The P value of GO terms was calculated (where n is the total number of genes, m the total number of genes that contain the term, c is the number of the genes of the list and k the genes of the list that contain the term). To study the functions of DEGs in progressive endometrial cancer, GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway [11] enrichment analysis were performed with hypergeometric distribution. P value less than 0.05 was set as the cut-off of GO terms and pathways.
Results
Results of preprocessing
After normalization of gene expression profile, intensity of genes in 8 samples were obtained, which was shown with box plot in Figure 1.
Identification of DEGs in progressive cancer
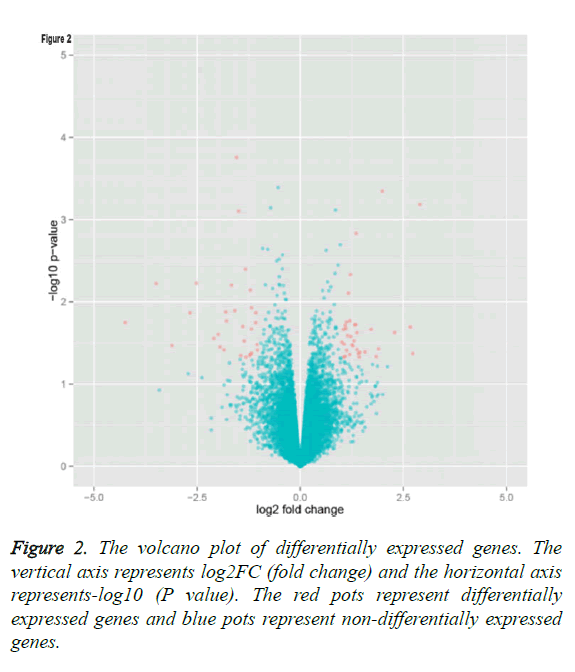
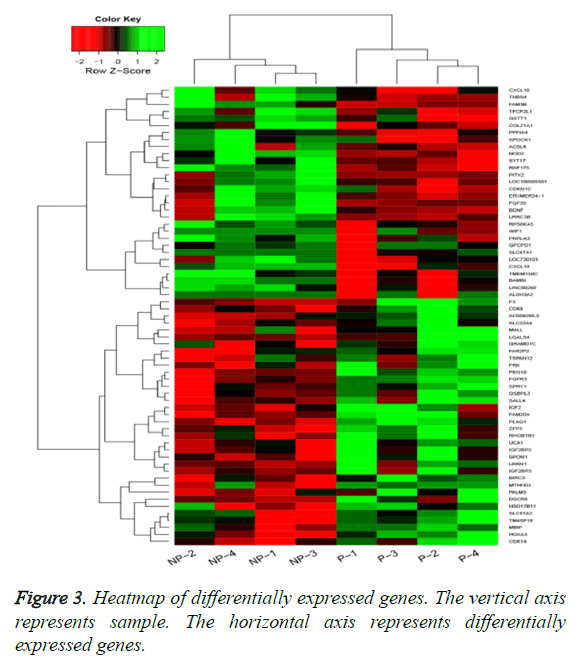
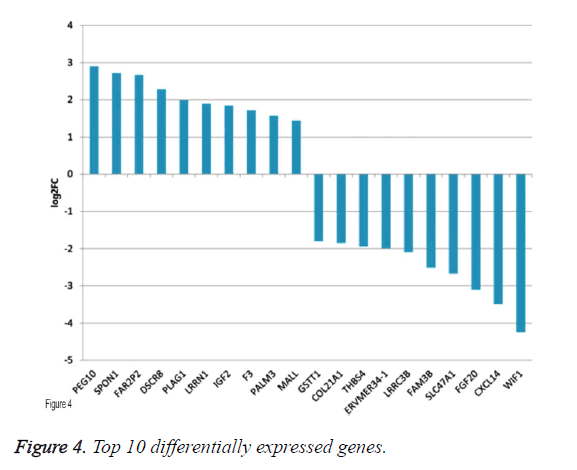
By applying statistical analysis, 65 DEGs were screened out between progressive and non-progressive samples, including 35 up- and 30 down-regulated genes. The volcano plot for DEGs, an effective and easy-to-interpret graph that summarizes both P value and fold-change, was shown in Figure 2. Results of hierarchical clustering were shown with heatmap, which indicated that progressive and non-progressive samples were clustered into two classes. The heatmap was shown in Figure 3. The top 10 up- and down-regulated DEGs were shown in Figure 4.
GO enrichment analysis of DEGs
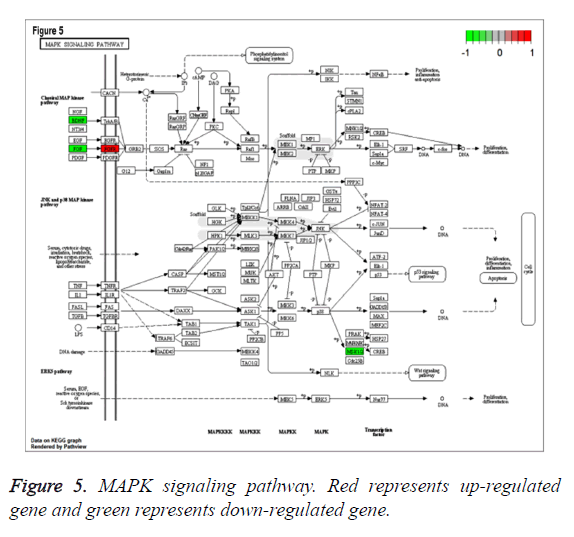
To study the functions of DEGs, GO enrichment analysis was performed and DEGs were mainly enriched in biological processes associated with cell proliferation which was involved up-regulated genes pleiomorphic adenoma gene 1 (PLAG1), insulin-like growth factor 2 (IGF2), coagulation factor III (F3), down-regulated genes fibroblast growth factor 20 (FGF20) and thrombospondin 4 (THBS4), categories of cell components including cytoplasmic cyclin-dependent protein kinase holoenzyme complex and categories of molecular functions such as mRNA 5’-UTR binding, protein kinase regulator activity (Table 1). Additionally, according to the criteria of P value less than 0.05, DEGs were significantly enriched in bladder cancer, fatty acid metabolism, MAPK signaling pathway involved with FGF20, glycerolipid metabolism, NOD-like receptor signaling pathway and TGF-beta signaling pathways involved with THBS4 (Table 2). Especially, MAPK signaling pathway, which was associated with most genes, was shown in Figure 5.
| GOBPID | Term | P value | Count | Symbols | |
|---|---|---|---|---|---|
| BP | GO: 0042127 | Regulation of cell proliferation | 1.50E-05 | 15 | BDNF; CDKN1C; F3; FGFR3; FRK; HOXA3; IGF2; CXCL10; PITX2; PLAG1; THBS4; SPRY1; BAMBI; FGF20; NOD2 |
| BP | GO: 0008284 | Positive regulation of cell proliferation | 2.63E-05 | 11 | F3; FGFR3; HOXA3; IGF2; CXCL10; PITX2; PLAG1; THBS4; BAMBI; FGF20; NOD2 |
| BP | GO: 0008283 | Cell proliferation | 9.11E-05 | 16 | BDNF; CDKN1C; F3; FGFR3; FRK; HOXA3; IGF2; CXCL10; PITX2; PLAG1; THBS4; SPRY1; BAMBI; FGF20; SALL4; NOD2 |
| BP | GO: 0007584 | Response to nutrient | 0.000197 | 5 | BDNF; ACSL4; CXCL10; PITX2; NOD2 |
| BP | GO: 0051239 | Regulation of multicellular organismal process | 0.000554 | 16 | BIRC3; BDNF; F3; FGFR3; CXCL10; PLAG1; SPOCK1; THBS4; RPS6KA5; SPRY1; IGF2BP3; IGF2BP2; TSPAN12; BAMBI; FGF20; NOD2 |
| BP | GO: 0009653 | Anatomical structure morphogenesis | 0.0006 | 17 | BDNF; F3; FGFR3; HOXA3; CXCL10; PITX2; PLAG1; THBS4; RPS6KA5; SPRY1; IGF2BP3; IGF2BP2; TSPAN12; BAMBI; TFCP2L1; FAM20A; SALL4 |
| BP | GO: 0050920 | Regulation of chemotaxis | 0.000657 | 4 | F3; CXCL10; THBS4; NOD2 |
| BP | GO: 0042490 | Mechanoreceptor differentiation | 0.000723 | 3 | BDNF; FGFR3; FGF20 |
| BP | GO: 0042325 | Regulation of phosphorylation | 0.000789 | 11 | BIRC3; CDKN1C; FGFR3; IGF2; CXCL10; THBS4; RPS6KA5; SPRY1; FGF20; MBIP; NOD2 |
| BP | GO: 0015695 | Organic cation transport | 0.000859 | 2 | SLC22A4; SLC47A1 |
| CC | GO: 0000308 | Cytoplasmic cyclin-dependent protein kinase holoenzyme complex | 0.006847 | 1 | CDK14 |
| CC | GO: 0009986 | Cell surface | 0.009575 | 6 | CD69; F3; FGFR3; CXCL10; PPFIA4; NOD2 |
| MF | GO: 0048027 | Mrna 5'-UTR binding | 0.00015 | 2 | IGF2BP3; IGF2BP2 |
| MF | GO: 0019887 | Protein kinase regulator activity | 0.000437 | 4 | CDKN1C; IGF2; CXCL10; MBIP |
| MF | GO: 0019207 | Kinase regulator activity | 0.000793 | 4 | CDKN1C; IGF2; CXCL10; MBIP |
| MF | GO: 0008083 | Growth factor activity | 0.001791 | 4 | BDNF; IGF2; THBS4; FGF20 |
| MF | GO: 0015491 | Cation: cation antiporter activity | 0.00224 | 2 | SLC22A4; SLC47A1 |
| MF | GO: 0005169 | Neurotrophin TRKB receptor binding | 0.003211 | 1 | BDNF |
| MF | GO: 0032500 | Muramyl dipeptide binding | 0.003211 | 1 | NOD2 |
| MF | GO: 0046577 | Long-chain-alcohol oxidase activity | 0.003211 | 1 | ALDH3A2 |
| MF | GO: 0047389 | Glycerophosphocholine phosphodiesterase activity | 0.003211 | 1 | GPCPD1 |
| MF | GO: 0047676 | Arachidonate-coa ligase activity | 0.003211 | 1 | ACSL4 |
Table 1: Results of Gene Ontology (GO) enrichment analysis of differentially expressed genes.
| KEGGID | Term | P value | Count | Symbols |
|---|---|---|---|---|
| hsa05219 | Bladder cancer | 0.007913 | 2 | FGFR3; RPS6KA5 |
| hsa00071 | Fatty acid metabolism | 0.008283 | 2 | ALDH3A2; ACSL4 |
| hsa04010 | MAPK signaling pathway | 0.009565 | 4 | BDNF; FGFR3; RPS6KA5; FGF20 |
| hsa00561 | Glycerolipid metabolism | 0.011086 | 2 | ALDH3A2; PNPLA3 |
| hsa04621 | NOD-like receptor signaling pathway | 0.014731 | 2 | BIRC3; NOD2 |
| hsa04350 | TGF-beta signaling pathway | 0.029556 | 2 | PITX2; THBS4 |
Table 2: Results of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of differentially expressed genes.
Discussion
Endometrial cancer spreads by exfoliation of cells that are shed through the fallopian tubes, by lymphatic and or hematogenous dissemination or by direct extension through the myometrium [12]. The most common route of endometrial cancer metastasis is the direct extension of the tumor cells to the myometrium, which results from several interdependent processes including breaking the basement membrane and extracellular matrix by proteolytic enzymes, thereby promoting cell invasion and distant metastasis [13,14].
In a previous study, a novel cytokine system which is composed of the receptor activator of nuclear factor- κB (RANK) and its ligand RANKL, has been indicated to be overexpressed in human endometrial cancer tissues [15]. Briefly, RANK/RANKL could promote cancer cell proliferation, migration and invasion. However, medroxyprogesterone acetate-mediated progesterone receptor B plays critical roles in inhibiting migration and invasion of endometrial cancer cells [15]. More recently, another gene alpha-enolase (ENO1), has been found to be up-regulated in several tumors [16]. Meanwhile, ENO1 silencing has obviously reduced cell proliferation, migration and invasion in endometrial cancer cells, as well as metastasis in vivo, as a result, it has been considered as a therapeutic target in treatment of endometrial carcinoma [17]. Additionally, a lysosomal cysteine protease, cathepsion B has also been overexpressed in endometrial cancer tissues and its silencing can inhibit cell migration and invasion [18]. Nevertheless, the molecular mechanism involved in the metastasis of endometrial cancer remains further elucidated.
In the present study, microarray analysis was performed to elucidate the molecular basis of progressive endometrial cancer and showed that 65 DEGs between progressive and nonprogressive cancer samples. From the results of functional annotation, we found that those genes were significantly enriched in GO terms of cell proliferation, including PLAG1, IGF2, F3, FGF20 and THBS4. Additionally, most of genes were involved in MAPK and TGF-beta signaling pathways.
PLAG1 is a proto-oncogene whose ectopic expression presumably results in the deregulation of target genes and leads to uncontrolled cell proliferation in tumorigenesis [19]. Studies have indicated that PLAG1 is involved in various human tumors including pleomorphic adenomas of the salivary glands [20], lipoblastomas [21], hepatoblastoma [22] and leukaemia [23]. Collectively, the PLAG1 proto-oncogene seems to exert its oncogenic potential at least partially via IGF2 mitogenic signaling pathway which leads to downstream signaling responsible for cell proliferation [24]. In our results, PLAG1 was up-regulated in progressive endometrial cancer samples and exerted important role in tumor cell proliferation, which was consistent with previous results indicated by other study. Furthermore, considering the critical function of IGF2 mitogenic signaling pathway, we also found that IGF2 was upregulated in progressive cancer samples and mainly involved in cell proliferation-associated functions. These findings were in line with results that increased plasma levels of IGF2 were associated with endometrial cancer risk [25].
It was also found that down-regulated genes FGF20 and THBS4 were not only associated with functions of cell proliferation, but also involved in MAPK and TGF-beta signaling pathways. FGF20 belongs to a large class of FGFs which encoding secretory proteins with potent mitogenic and angiogenic roles in tumor development [26]. Previous study has shown that FGF20 was expressed at markedly elevated levels in part of human ovarian endometrioid adenocarcinomas harboring β-catenin defects [27]. However, FGF20 was downregulated in progressive cancer sample, and one possible explanation is that the gene was under the control of more than one pathway. Notably, FGF20 has been proved to activate the MAPK signaling pathway which is the major intracellular signaling pathway of FGFs and plays a crucial role in the neurotrophic activity of FGF20 [28]. Consistently, our bioinformatics results indicated that FGF20 exerted crucial role in MAPK signaling pathway. THBS4, a member of the extracellular calcium-binding protein family, is involved in cell adhesion and migration, and considered as a putative tumor suppressor gene in tumorigenesis [29,30]. In our study, expression levels of THBS4 were significantly decreased in progressive samples compared with non-progressive control. Moreover, THBS4 was involved in TGF-beta signaling pathway, which acted as a suppressor of epithelial cell tumorigenesis at early stages, but promoted cancer progression by contributing to tumor cell migration and invasion during the later stages. Taken together, these results suggest these genes have metastatic potential in endometrial cancer.
In conclusion, our results showed that 65 genes were differentially expressed in progressive endometrial cancer samples compared with non-progressive control, which were mainly associated with tumor metastasis possibly by enhancing cell proliferation and affecting MAPK and TGF-beta signaling pathways. High expression of PLAG1, IGF2 and low expression of FGF20, THBS4, therefore, appear to play an important role in tumorigenesis and progression of endometrial cancer. However, these findings warrant further prospective studies to be validated and determine if these genes could serve as new molecular targets for endometrial cancer therapy.
Acknowledgment
None
Conflict of Interest
We certify that regarding this paper, no actual or potential conflicts of interests exist.
References
- Torre LA. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87-108.
- Bidus MA. Prediction of lymph node metastasis in patients with endometrioid endometrial cancer using expression microarray. Clin Cancer Res 2006; 12: 83-88.
- Balch C. Role of epigenomics in ovarian and endometrial cancers. Epigenomics 2010; 2: 419-447.
- Amant F. Endometrial cancer. Lancet 2005; 366: 491-505.
- Colombo N. Endometrial cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2011; 22: 35-39.
- Gautier L. Affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004; 20: 307-315.
- Irizarry RA. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003; 4: 249-264.
- Smyth GK. Limma: linear models for microarray data, in Bioinformatics and computational biology solutions using R and bioconductor. Springer 2005; 397-420.
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007; 8: 118-127.
- Ashburner M. Gene ontology: tool for the unification of biology. Nat Gene 2000; 25: 25-29.
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucl Acids Res 2000; 28: 27-30.
- Sato R, Jobo T, Kuramoto H. Parametrial spread is a prognostic factor in endometrial carcinoma. Eur J Gynaecol Oncol 2002; 24: 241-245.
- Jedinak A, Maliar T. Inhibitors of proteases as anticancer drugs. Neoplasma 2004; 52: 185-192.
- Hornebeck W. Matrix-directed regulation of pericellular proteolysis and tumor progression. Sem Cancer Biol 2002.
- Wang J. MPA influences tumor cell proliferation, migration, and invasion induced by RANKL through PRB involving the MAPK pathway in endometrial cancer. Oncol Rep 2015; 33: 799-809.
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer 2011; 11: 85-95.
- Zhao M. Enolase-1 is a therapeutic target in endometrial carcinoma. Oncotarget 2015.
- Bao W. Silencing of cathepsin B suppresses the proliferation and invasion of endometrial cancer. Oncol Rep 2013; 30: 723-730.
- Voz ML. Microarray screening for target genes of the proto-oncogene PLAG1. Oncogene 2004; 23: 179-191.
- Asp J. CHCHD7‐PLAG1 and TCEA1‐PLAG1 gene fusions resulting from cryptic, intrachromosomal 8q rearrangements in pleomorphic salivary gland adenomas. Genes Chromos Cancer 2006; 45: 820-828.
- Astrom A. Evidence of involvement of the PLAG1 gene in lipoblastomas. Int J Oncol 2000; 16: 1107-1117.
- Zatkova A. Amplification and overexpression of the IGF2 regulator PLAG1 in hepatoblastoma. Genes Chromos Cancer 2004; 39: 126-137.
- Castilla LH. Identification of genes that synergize with Cbfb-MYH11 in the pathogenesis of acute myeloid leukemia. Proc Nat Acad Sci USA 2004; 101: 4924-4929.
- Samani AA. The role of the IGF system in cancer growth and metastasis: overview and recent insights. Endocrine Rev 2007; 28: 20-47.
- Oh JC. Increased plasma levels of insulin-like growth factor 2 and insulin-like growth factor binding protein 3 are associated with endometrial cancer risk. Cancer Epidemiol Biomark Prev 2004; 13: 748-752.
- Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 2010; 10: 116-129.
- Chamorro MN. FGF‐20 and DKK1 are transcriptional targets of β-catenin and FGF‐20 is implicated in cancer and development. EMBO J 2005; 24: 73-84.
- Ohmachi S. Preferential neurotrophic activity of fibroblast growth factor‐20 for dopaminergic neurons through fibroblast growth factor receptor‐1c. J Neurosci Res 2003; 72: 436-443.
- Korkola JE. Differentiation of lobular versus ductal breast carcinomas by expression microarray analysis. Cancer Res 2003; 63: 7167-7175.
- van Doorn R. Epigenetic profiling of cutaneous T-cell lymphoma: promoter hypermethylation of multiple tumor suppressor genes including BCL7a, PTPRG, and p73. J Clin Oncol 2005; 23: 3886-3896.